Spatiotemporal Imaging of Cellular Energy Metabolism with Genetically-Encoded Fluorescent Sensors in Brain
- PMID: 29679217
- PMCID: PMC6129245
- DOI: 10.1007/s12264-018-0229-3
Spatiotemporal Imaging of Cellular Energy Metabolism with Genetically-Encoded Fluorescent Sensors in Brain
Abstract
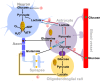
The brain has very high energy requirements and consumes 20% of the oxygen and 25% of the glucose in the human body. Therefore, the molecular mechanism underlying how the brain metabolizes substances to support neural activity is a fundamental issue for neuroscience studies. A well-known model in the brain, the astrocyte-neuron lactate shuttle, postulates that glucose uptake and glycolytic activity are enhanced in astrocytes upon neuronal activation and that astrocytes transport lactate into neurons to fulfill their energy requirements. Current evidence for this hypothesis has yet to reach a clear consensus, and new concepts beyond the shuttle hypothesis are emerging. The discrepancy is largely attributed to the lack of a critical method for real-time monitoring of metabolic dynamics at cellular resolution. Recent advances in fluorescent protein-based sensors allow the generation of a sensitive, specific, real-time readout of subcellular metabolites and fill the current technological gap. Here, we summarize the development of genetically encoded metabolite sensors and their applications in assessing cell metabolism in living cells and in vivo, and we believe that these tools will help to address the issue of elucidating neural energy metabolism.
Keywords: Astrocyte; Energy metabolism; Genetically encoded fluorescent sensor; Neuron; Real time monitoring.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

