Light availability impacts structure and function of phototrophic stream biofilms across domains and trophic levels
- PMID: 29679511
- PMCID: PMC6055792
- DOI: 10.1111/mec.14696
Light availability impacts structure and function of phototrophic stream biofilms across domains and trophic levels
Abstract
Phototrophic biofilms are ubiquitous in freshwater and marine environments where they are critical for biogeochemical cycling, food webs and in industrial applications. In streams, phototrophic biofilms dominate benthic microbial life and harbour an immense prokaryotic and eukaryotic microbial biodiversity with biotic interactions across domains and trophic levels. Here, we examine how community structure and function of these biofilms respond to varying light availability, as the crucial energy source for phototrophic biofilms. Using metatranscriptomics, we found that under light limitation-dominant phototrophs, including diatoms and cyanobacteria, displayed a remarkable plasticity in their photosynthetic machinery manifested as higher abundance of messenger RNAs (mRNAs) involved in photosynthesis and chloroplast ribosomal RNA. Under higher light availability, bacterial mRNAs involved in phosphorus metabolism, mainly from Betaproteobacteria and Cyanobacteria, increased, likely compensating for nutrient depletion in thick biofilms with high biomass. Consumers, including diverse ciliates, displayed community shifts indicating preferential grazing on algae instead of bacteria under higher light. For the first time, we show that the functional integrity of stream biofilms under variable light availability is maintained by structure-function adaptations on several trophic levels. Our findings shed new light on complex biofilms, or "microbial jungles", where in analogy to forests, diverse and multitrophic level communities lend stability to ecosystem functioning. This multitrophic level perspective, coupling metatranscriptomics to process measurements, could advance understanding of microbial-driven ecosystems beyond biofilms, including planktonic and soil environments.
Keywords: rRNA; diatoms; freshwater; mRNA; metatranscriptomics; microbial eukaryotes.
© 2018 The Authors. Molecular Ecology Published by John Wiley & Sons Ltd.
Figures

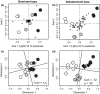
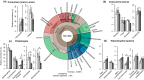

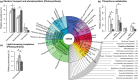
Comment in
-
Testing the light:nutrient hypothesis: Insights into biofilm structure and function using metatranscriptomics.Mol Ecol. 2018 Jul;27(14):2909-2912. doi: 10.1111/mec.14733. Mol Ecol. 2018. PMID: 29998558
References
-
- Adlboller, M. (2013). Hydrologic stochasticity and biodiversity of biofilm algae. MSc Thesis, University of Vienna.
-
- Alexander, H. , Jenkins, B. D. , Rynearson, T. A. , & Dyhrman, S. T. (2015). Metatranscriptome analyses indicate resource partitioning between diatoms in the field. Proceedings of the National Academy of Sciences of the United States of America, 112, 2182–2190. 10.1073/pnas.1421993112 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

