A phylogenetic framework facilitates Y-STR variant discovery and classification via massively parallel sequencing
- PMID: 29679929
- PMCID: PMC6010625
- DOI: 10.1016/j.fsigen.2018.03.012
A phylogenetic framework facilitates Y-STR variant discovery and classification via massively parallel sequencing
Abstract
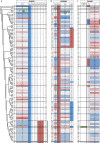
Short tandem repeats on the male-specific region of the Y chromosome (Y-STRs) are permanently linked as haplotypes, and therefore Y-STR sequence diversity can be considered within the robust framework of a phylogeny of haplogroups defined by single nucleotide polymorphisms (SNPs). Here we use massively parallel sequencing (MPS) to analyse the 23 Y-STRs in Promega's prototype PowerSeq™ Auto/Mito/Y System kit (containing the markers of the PowerPlex® Y23 [PPY23] System) in a set of 100 diverse Y chromosomes whose phylogenetic relationships are known from previous megabase-scale resequencing. Including allele duplications and alleles resulting from likely somatic mutation, we characterised 2311 alleles, demonstrating 99.83% concordance with capillary electrophoresis (CE) data on the same sample set. The set contains 267 distinct sequence-based alleles (an increase of 58% compared to the 169 detectable by CE), including 60 novel Y-STR variants phased with their flanking sequences which have not been reported previously to our knowledge. Variation includes 46 distinct alleles containing non-reference variants of SNPs/indels in both repeat and flanking regions, and 145 distinct alleles containing repeat pattern variants (RPV). For DYS385a,b, DYS481 and DYS390 we observed repeat count variation in short flanking segments previously considered invariable, and suggest new MPS-based structural designations based on these. We considered the observed variation in the context of the Y phylogeny: several specific haplogroup associations were observed for SNPs and indels, reflecting the low mutation rates of such variant types; however, RPVs showed less phylogenetic coherence and more recurrence, reflecting their relatively high mutation rates. In conclusion, our study reveals considerable additional diversity at the Y-STRs of the PPY23 set via MPS analysis, demonstrates high concordance with CE data, facilitates nomenclature standardisation, and places Y-STR sequence variants in their phylogenetic context.
Keywords: Massively parallel sequencing; PPY23; PowerSeq system; Repeat pattern variation (RPV); Single nucleotide polymorphism (SNP); Y-STRs.
Copyright © 2018 The Authors. Published by Elsevier B.V. All rights reserved.
Figures

Similar articles
-
Investigation into the sequence structure of 23 Y chromosomal STR loci using massively parallel sequencing.Forensic Sci Int Genet. 2016 Nov;25:132-141. doi: 10.1016/j.fsigen.2016.08.010. Epub 2016 Aug 28. Forensic Sci Int Genet. 2016. PMID: 27591816
-
Sequencing of autosomal, mitochondrial and Y-chromosomal forensic markers in the People of the British Isles cohort detects population structure dominated by patrilineages.Forensic Sci Int Genet. 2022 Jul;59:102725. doi: 10.1016/j.fsigen.2022.102725. Epub 2022 May 18. Forensic Sci Int Genet. 2022. PMID: 35640311
-
Genetic analysis of Southern Brazil subjects using the PowerSeq™ AUTO/Y system for short tandem repeat sequencing.Forensic Sci Int Genet. 2018 Mar;33:129-135. doi: 10.1016/j.fsigen.2017.12.008. Epub 2017 Dec 16. Forensic Sci Int Genet. 2018. PMID: 29275088
-
Forensic molecular biomarkers for mixture analysis.Forensic Sci Int Genet. 2019 Jul;41:107-119. doi: 10.1016/j.fsigen.2019.04.003. Epub 2019 Apr 30. Forensic Sci Int Genet. 2019. PMID: 31071519 Review.
-
Current state-of-art of STR sequencing in forensic genetics.Electrophoresis. 2018 Nov;39(21):2655-2668. doi: 10.1002/elps.201800030. Epub 2018 Jun 4. Electrophoresis. 2018. PMID: 29750373 Review.
Cited by
-
On the Forensic Use of Y-Chromosome Polymorphisms.Genes (Basel). 2022 May 17;13(5):898. doi: 10.3390/genes13050898. Genes (Basel). 2022. PMID: 35627283 Free PMC article. Review.
-
Sequencing of human identification markers in an Uyghur population using the MiSeq FGxTM Forensic Genomics System.Forensic Sci Res. 2020 Sep 10;7(2):154-162. doi: 10.1080/20961790.2020.1779967. eCollection 2022. Forensic Sci Res. 2020. PMID: 35784409 Free PMC article.
-
Massively parallel sequencing and capillary electrophoresis of a novel panel of falcon STRs: Concordance with minisatellite DNA profiles from historical wildlife crime.Forensic Sci Int Genet. 2021 Sep;54:102550. doi: 10.1016/j.fsigen.2021.102550. Epub 2021 Jun 10. Forensic Sci Int Genet. 2021. PMID: 34174583 Free PMC article.
-
A multi-dimensional evaluation of the 'NIST 1032' sample set across four forensic Y-STR multiplexes.Forensic Sci Int Genet. 2022 Mar;57:102655. doi: 10.1016/j.fsigen.2021.102655. Epub 2021 Dec 28. Forensic Sci Int Genet. 2022. PMID: 35007854 Free PMC article.
-
Sequencing the orthologs of human autosomal forensic short tandem repeats provides individual- and species-level identification in African great apes.BMC Ecol Evol. 2024 Oct 31;24(1):134. doi: 10.1186/s12862-024-02324-0. BMC Ecol Evol. 2024. PMID: 39482599 Free PMC article.
References
-
- Butler J.M. Academic Press; Cambridge MA: 2009. Fundamentals of Forensic DNA Typing.
-
- Weber J.L., Wong C. Mutation of human short tandem repeats. Hum. Mol. Genet. 1993;3:1123–1128. - PubMed
-
- Jobling M.A., Pandya A., Tyler-Smith C. The Y chromosome in forensic analysis and paternity testing. Int. J. Legal Med. 1997;110:118–124. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

