The Dystrophin Glycoprotein Complex Regulates the Epigenetic Activation of Muscle Stem Cell Commitment
- PMID: 29681515
- PMCID: PMC5935555
- DOI: 10.1016/j.stem.2018.03.022
The Dystrophin Glycoprotein Complex Regulates the Epigenetic Activation of Muscle Stem Cell Commitment
Abstract
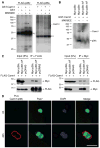
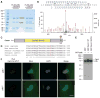
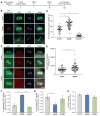
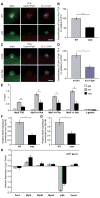
Asymmetrically dividing muscle stem cells in skeletal muscle give rise to committed cells, where the myogenic determination factor Myf5 is transcriptionally activated by Pax7. This activation is dependent on Carm1, which methylates Pax7 on multiple arginine residues, to recruit the ASH2L:MLL1/2:WDR5:RBBP5 histone methyltransferase complex to the proximal promoter of Myf5. Here, we found that Carm1 is a specific substrate of p38γ/MAPK12 and that phosphorylation of Carm1 prevents its nuclear translocation. Basal localization of the p38γ/p-Carm1 complex in muscle stem cells occurs via binding to the dystrophin-glycoprotein complex (DGC) through β1-syntrophin. In dystrophin-deficient muscle stem cells undergoing asymmetric division, p38γ/β1-syntrophin interactions are abrogated, resulting in enhanced Carm1 phosphorylation. The resulting progenitors exhibit reduced Carm1 binding to Pax7, reduced H3K4-methylation of chromatin, and reduced transcription of Myf5 and other Pax7 target genes. Therefore, our experiments suggest that dysregulation of p38γ/Carm1 results in altered epigenetic gene regulation in Duchenne muscular dystrophy.
Keywords: Carm1; Duchenne muscular dystrophy; Pax7; asymmetric division; cell polarity; muscle stem cell; p38γ MAPK; satellite cell; stem cell expansion; β1-syntrophin.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Campbell KP, Kahl SD. Association of dystrophin and an integral membrane glycoprotein. Nature. 1989;338:259–262. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

