Efficient Gene Transfer and Gene Editing in Sterlet (Acipenser ruthenus)
- PMID: 29681919
- PMCID: PMC5897424
- DOI: 10.3389/fgene.2018.00117
Efficient Gene Transfer and Gene Editing in Sterlet (Acipenser ruthenus)
Abstract
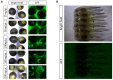
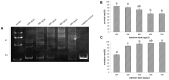
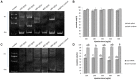
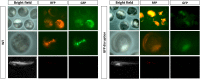
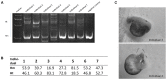
The sturgeon (Acipenseriformes) is an important farmed species because of its economical value. However, neither gene transfer nor gene editing techniques have been established in sturgeon for molecular breeding and gene functional study until now. In this study, we accomplished gene transfer and gene editing in sterlet (Acipenser ruthenus), which has the shortest sexual maturation period of sturgeons. The plasmid encoding enhanced green fluorescent protein (EGFP) was transferred into the embryos of sterlet at injection concentration of 100 ng/μL, under which condition high survival rate and gene transfer rate could be achieved. Subsequently, exogenous EGFP was efficiently disrupted by transcription activator-like effector nucleases (TALENs) or clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 nuclease/guide RNA (gRNA), with injection concentrations of 300 ng/μL TALENs, or 100 ng/μL Cas9 nuclease and 30 ng/μL gRNA, respectively, under which condition high survival rate and gene mutation rate could be achieved. Finally, the endogenous gene no tail in sterlet was successfully mutated by Cas9 nuclease/gRNA. We observed the CRISPR-induced no tail mutation, at a high efficiency with the mutant P0 embryos displaying the expected phenotype of bent spine and twisted tail.
Keywords: EGFP; gene editing; gene transfer; no tail; sterlet.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

