Glycogen debranching enzyme (AGL) is a novel regulator of non-small cell lung cancer growth
- PMID: 29682180
- PMCID: PMC5908281
- DOI: 10.18632/oncotarget.24676
Glycogen debranching enzyme (AGL) is a novel regulator of non-small cell lung cancer growth
Abstract
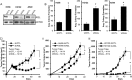
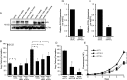
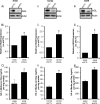
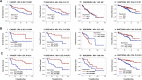
Glycogen debranching enzyme (AGL) and Glycogen phosphorylase (PYG) are responsible for glycogen breakdown. We have earlier shown that AGL is a regulator of bladder tumor growth. Here we investigate the role of AGL in non-small cell lung cancers (NSCLC). Short hairpin RNA (shRNA) driven knockdown of AGL resulted in increased anchorage independent and xenograft growth of NSCLC cells. We further establish that an increase in hyaluronic acid (HA) synthesis driven by Hyaluronic Acid Synthase 2 (HAS2) is critical for anchorage independent growth of NSCLC cells with AGL loss. Using gene knockdown approach against HAS2 and by using 4-methylumbelliferone (4MU), an inhibitor of HA synthesis, we show that HA synthesis is critical for growth of NSCLC cells that have lost AGL. We further show NSCLC cells without AGL expression are dependent on RHAMM for HA signaling and growth. Analysis of NSCLC patient datasets established that patients with low AGL/high HAS2 or low AGL/high RHAMM mRNA expression have poor overall survival compared to patients with high AGL/low HAS2 or high AGL/low RHAMM expression. We show for the first time that loss of AGL promotes anchorage independent growth of NSCLC cells. We further show that HAS2 driven HA synthesis and signaling via RHAMM is critical in regulating growth of these cancer cells with AGL loss. Further patients presenting with low AGL and HAS2 or RHAMM over expressing tumors might present the ideal cohort who would respond to inhibitors of HA synthesis and signaling.
Keywords: AGL; HAS2; RHAMM; hyaluronic acid; non-small cell lung cancer.
Conflict of interest statement
CONFLICTS OF INTEREST The authors declare no conflicts of interest.
Figures



Similar articles
-
CD44 and RHAMM are essential for rapid growth of bladder cancer driven by loss of Glycogen Debranching Enzyme (AGL).BMC Cancer. 2016 Sep 5;16(1):713. doi: 10.1186/s12885-016-2756-5. BMC Cancer. 2016. PMID: 27595989 Free PMC article.
-
Loss of Glycogen Debranching Enzyme AGL Drives Bladder Tumor Growth via Induction of Hyaluronic Acid Synthesis.Clin Cancer Res. 2016 Mar 1;22(5):1274-83. doi: 10.1158/1078-0432.CCR-15-1706. Epub 2015 Oct 21. Clin Cancer Res. 2016. PMID: 26490312 Free PMC article.
-
Involvement of glycogen debranching enzyme in bladder cancer.Biomed Rep. 2017 Jun;6(6):595-598. doi: 10.3892/br.2017.907. Epub 2017 May 9. Biomed Rep. 2017. PMID: 28584628 Free PMC article.
-
Targeting glycogen metabolism in bladder cancer.Nat Rev Urol. 2015 Jul;12(7):383-91. doi: 10.1038/nrurol.2015.111. Epub 2015 May 26. Nat Rev Urol. 2015. PMID: 26032551 Free PMC article. Review.
-
Metabolic phenotype of bladder cancer.Cancer Treat Rev. 2016 Apr;45:46-57. doi: 10.1016/j.ctrv.2016.03.005. Epub 2016 Mar 8. Cancer Treat Rev. 2016. PMID: 26975021 Review.
Cited by
-
Y chromosome loss in cancer drives growth by evasion of adaptive immunity.Nature. 2023 Jul;619(7970):624-631. doi: 10.1038/s41586-023-06234-x. Epub 2023 Jun 21. Nature. 2023. PMID: 37344596 Free PMC article.
-
Genome-Wide DNA Methylation Confirms Oral Squamous Cell Carcinomas in Proliferative Verrucous Leukoplakia as a Distinct Oral Cancer Subtype: A Case-Control Study.Cancers (Basel). 2025 Jan 13;17(2):245. doi: 10.3390/cancers17020245. Cancers (Basel). 2025. PMID: 39858027 Free PMC article.
-
Molecular subtyping of esophageal squamous cell carcinoma by large-scale transcriptional profiling: Characterization, therapeutic targets, and prognostic value.Front Genet. 2022 Nov 8;13:1033214. doi: 10.3389/fgene.2022.1033214. eCollection 2022. Front Genet. 2022. PMID: 36425064 Free PMC article.
-
Revisiting Glycogen in Cancer: A Conspicuous and Targetable Enabler of Malignant Transformation.Front Oncol. 2020 Oct 30;10:592455. doi: 10.3389/fonc.2020.592455. eCollection 2020. Front Oncol. 2020. PMID: 33224887 Free PMC article. Review.
-
CD44 in Bladder Cancer.Cancers (Basel). 2024 Mar 18;16(6):1195. doi: 10.3390/cancers16061195. Cancers (Basel). 2024. PMID: 38539529 Free PMC article. Review.
References
-
- Murray RK, Granner DK, Mayes PA, Rodwell VW. Harper2019;s Illustrated Biochemistry. McGraw Hill; 2003.
-
- Kishnani PS, Austin SL, Arn P, Bali DS, Boney A, Case LE, Chung WK, Desai DM, El-Gharbawy A, Haller R, Smit GP, Smith AD, Hobson-Webb LD, et al. ACMG Glycogen storage disease type III diagnosis and management guidelines. Genet Med. 2010;12:446–63. https://doi.org/10.1097/GIM.0b013e3181e655b600125817-201007000-00008 - DOI - PubMed
-
- Guin S, Pollard C, Ru Y, Ritterson Lew C, Duex JE, Dancik G, Owens C, Spencer A, Knight S, Holemon H, Gupta S, Hansel D, Hellerstein M, et al. Role in tumor growth of a glycogen debranching enzyme lost in glycogen storage disease. J Natl Cancer Inst. 2014;106:dju062. https://doi.org/10.1093/jnci/dju062 - DOI - PMC - PubMed
-
- Guin S, Ru Y, Agarwal N, Lew CR, Owens C, Comi GP, Theodorescu D. Loss of Glycogen Debranching Enzyme AGL Drives Bladder Tumor Growth via Induction of Hyaluronic Acid Synthesis. Clin Cancer Res. 2016;22:1274–83. https://doi.org/10.1158/1078-0432.CCR-15-1706 - DOI - PMC - PubMed
-
- Oldenburg D, Ru Y, Weinhaus B, Cash S, Theodorescu D, Guin S. CD44 and RHAMM are essential for rapid growth of bladder cancer driven by loss of Glycogen Debranching Enzyme (AGL) BMC Cancer. 2016;16:713. https://doi.org/10.1186/s12885-016-2756-5 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

