DNA-encoded chemical libraries - achievements and remaining challenges
- PMID: 29683493
- PMCID: PMC6126621
- DOI: 10.1002/1873-3468.13068
DNA-encoded chemical libraries - achievements and remaining challenges
Abstract
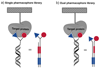
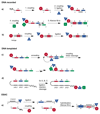
DNA-encoded chemical libraries (DECLs) are collections of compounds, individually coupled to DNA tags serving as amplifiable identification barcodes. Since individual compounds can be identified by the associated DNA tag, they can be stored as a mixture, allowing the synthesis and screening of combinatorial libraries of unprecedented size, facilitated by the implementation of split-and-pool synthetic procedures or other experimental methodologies. In this review, we briefly present relevant concepts and technologies, which are required for the implementation and interpretation of screening procedures with DNA-encoded chemical libraries. Moreover, we illustrate some success stories, detailing how novel ligands were discovered from encoded libraries. Finally, we critically review what can realistically be achieved with the technology at the present time, highlighting challenges and opportunities for the future.
Keywords: DNA; DNA-encoded chemical libraries; combinatorial chemistry; drug discovery; high-throughput DNA sequencing.
© 2018 Federation of European Biochemical Societies.
Figures
Similar articles
-
DNA-encoded chemical libraries: advancing beyond conventional small-molecule libraries.Acc Chem Res. 2014 Apr 15;47(4):1247-55. doi: 10.1021/ar400284t. Epub 2014 Mar 28. Acc Chem Res. 2014. PMID: 24673190
-
On-DNA hit validation methodologies for ligands identified from DNA-encoded chemical libraries.Biochem Biophys Res Commun. 2020 Dec 3;533(2):235-240. doi: 10.1016/j.bbrc.2020.04.030. Epub 2020 Apr 30. Biochem Biophys Res Commun. 2020. PMID: 32362331
-
DNA-Encoded Chemical Libraries: A Selection System Based on Endowing Organic Compounds with Amplifiable Information.Annu Rev Biochem. 2018 Jun 20;87:479-502. doi: 10.1146/annurev-biochem-062917-012550. Epub 2018 Jan 12. Annu Rev Biochem. 2018. PMID: 29328784 Free PMC article. Review.
-
20 years of DNA-encoded chemical libraries.Chem Commun (Camb). 2011 Dec 28;47(48):12747-53. doi: 10.1039/c1cc15634a. Epub 2011 Nov 14. Chem Commun (Camb). 2011. PMID: 22083211 Review.
-
Achievements, Challenges, and Opportunities in DNA-Encoded Library Research: An Academic Point of View.Chembiochem. 2017 May 4;18(9):829-836. doi: 10.1002/cbic.201600567. Epub 2017 Jan 25. Chembiochem. 2017. PMID: 28032411 Review.
Cited by
-
Universal encoding of next generation DNA-encoded chemical libraries.Chem Sci. 2022 Jan 11;13(4):967-974. doi: 10.1039/d1sc05721a. eCollection 2022 Jan 26. Chem Sci. 2022. PMID: 35211261 Free PMC article.
-
Comparative evaluation of DNA-encoded chemical selections performed using DNA in single-stranded or double-stranded format.Biochem Biophys Res Commun. 2020 Dec 3;533(2):223-229. doi: 10.1016/j.bbrc.2020.04.035. Epub 2020 May 5. Biochem Biophys Res Commun. 2020. PMID: 32386812 Free PMC article.
-
Design and synthesis of a chemically diverse, lead-like DNA-encoded library from sequential amide coupling.RSC Med Chem. 2025 Jul 29. doi: 10.1039/d5md00350d. Online ahead of print. RSC Med Chem. 2025. PMID: 40786536 Free PMC article.
-
Negishi-coupling-enabled synthesis of α-heteroaryl-α-amino acid building blocks for DNA-encoded chemical library applications.Beilstein J Org Chem. 2024 Aug 8;20:1922-1932. doi: 10.3762/bjoc.20.168. eCollection 2024. Beilstein J Org Chem. 2024. PMID: 39135657 Free PMC article.
-
Homogeneous and Functional Group Tolerant Ring-Closing Metathesis for DNA-Encoded Chemical Libraries.ACS Comb Sci. 2020 Feb 10;22(2):80-88. doi: 10.1021/acscombsci.9b00199. Epub 2020 Jan 21. ACS Comb Sci. 2020. PMID: 31913011 Free PMC article.
References
-
- Goodnow RA, Jr, Dumelin CE, Keefe AD. DNA-encoded chemistry: enabling the deeper sampling of chemical space. Nat Rev Drug Discov. 2017;16:131–147. - PubMed
-
- McCafferty J, Griffiths AG, W G, Chiswell D. Phage antibodies: filamentous phage displaying antibody variable domains. Nature. 1990;348:3. - PubMed
-
- Winter G, Milstein C. Man-made antibodies. Nature. 1991;349:7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

