Neutral Theory and the Somatic Evolution of Cancer
- PMID: 29684198
- PMCID: PMC5967571
- DOI: 10.1093/molbev/msy079
Neutral Theory and the Somatic Evolution of Cancer
Erratum in
-
Neutral Theory and the Somatic Evolution of Cancer.Mol Biol Evol. 2018 Jun 1;35(6):1561. doi: 10.1093/molbev/msy100. Mol Biol Evol. 2018. PMID: 29800319 Free PMC article. No abstract available.
Abstract
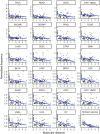
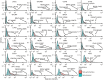
Kimura's neutral theory argued that positive selection was not responsible for an appreciable fraction of molecular substitutions. Correspondingly, quantitative analysis reveals that the vast majority of substitutions in cancer genomes are not detectably under selection. Insights from the somatic evolution of cancer reveal that beneficial substitutions in cancer constitute a small but important fraction of the molecular variants. The molecular evolution of cancer community will benefit by incorporating the neutral theory of molecular evolution into their understanding and analysis of cancer evolution-and accepting the use of tractable, predictive models, even when there is some evidence that they are not perfect.
Figures
References
-
- Altrock PM, Liu LL, Michor F.. 2015. The mathematics of cancer: integrating quantitative models. Nat Rev Cancer. 15(12):730–745. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

