The role of natural selection in shaping genetic variation in a promising Chagas disease drug target: Trypanosoma cruzi trans-sialidase
- PMID: 29684709
- PMCID: PMC6196115
- DOI: 10.1016/j.meegid.2018.04.025
The role of natural selection in shaping genetic variation in a promising Chagas disease drug target: Trypanosoma cruzi trans-sialidase
Abstract
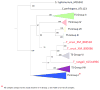
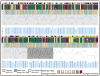
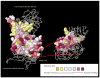
Rational drug design creates innovative therapeutics based on knowledge of the biological target to provide more effective and responsible therapeutics. Chagas disease, endemic throughout Latin America, is caused by Trypanosoma cruzi, a protozoan parasite. Current therapeutics are problematic with widespread calls for new approaches. Researchers are using rational drug design for Chagas disease and one target receiving considerable attention is the T. cruzi trans-sialidase protein (TcTS). In T. cruzi, trans-sialidase catalyzes the transfer of sialic acid from a mammalian host to coat the parasite surface membrane and avoid immuno-detection. However, the role of TcTS in pathology variance among and within genetic variants of the parasite is not well understood despite numerous studies. Previous studies reported the crystalline structure of TcTS and the TS protein structure in other trypanosomes where the enzyme is often inactive. However, no study has examined the role of natural selection in genetic variation in TcTS. To understand the role of natural selection in TcTS DNA sequence and protein variation, we examined a 471 bp portion of the TcTS gene from 48 T. cruzi samples isolated from insect vectors. Because there may be multiple parasite genotypes infecting one insect and there are multiple copies of TcTS per parasite genome, all 48 sequences had multiple polymorphic bases. To resolve these polymorphisms, we examined cloned sequences from two insect vectors. The data are analyzed to understand the role of natural selection in shaping genetic variation in TcTS and interpreted in light of the possible role of TcTS as a drug target. The analysis highlights negative or purifying selection on three amino acids previously shown to be important in TcTS transfer activity. One amino acid in particular, Tyr342, is a strong candidate for a drug target because it is under negative selection and amino acid substitutions inactivate TcTS transfer activity. AUTHOR SUMMARY: Chagas disease is caused by the protozoan parasite Trypanosoma cruzi and transmitted to humans and other mammals primarily by Triatomine insects. Being endemic in many South and Central American countries and affecting millions of people the need for new more effective and safe therapies is evident. Here, we examine genetic variation and natural selection on DNA (471 bp) and amino acid (157 aa) sequence data of the T. cruzi trans-sialdiase (TcTS) protein, often suggested as a candidate for rational drug design. In our surveyed region of the protein there were five amino acid residues that have been shown to be integral to the function of TcTS. We found that three were under strong negative selection making them ideal candidates for drug design; however, one was under balancing selection and should be avoided as a drug target. Our study provides new information into identifying potential targets for a new Chagas drug.
Keywords: Chagas disease; Genetic variation; Natural selection; Rational drug design; Trans-sialidase; Triatoma dimidiata; Triatoma nitida; Trypanosoma cruzi.
Copyright © 2018 Elsevier B.V. All rights reserved.
Figures


Similar articles
-
Recombination-driven generation of the largest pathogen repository of antigen variants in the protozoan Trypanosoma cruzi.BMC Genomics. 2016 Sep 13;17(1):729. doi: 10.1186/s12864-016-3037-z. BMC Genomics. 2016. PMID: 27619017 Free PMC article.
-
Rational drug design in parasitology: trans-sialidase as a case study for Chagas disease.Drug Discov Today. 2008 Feb;13(3-4):110-7. doi: 10.1016/j.drudis.2007.12.004. Drug Discov Today. 2008. PMID: 18275908 Review.
-
Design of e-pharmacophore models using compound fragments for the trans-sialidase of Trypanosoma cruzi: screening for novel inhibitor scaffolds.J Mol Graph Model. 2013 Sep;45:84-97. doi: 10.1016/j.jmgm.2013.08.009. Epub 2013 Aug 16. J Mol Graph Model. 2013. PMID: 24012872 Free PMC article.
-
Synthesis, molecular docking and biological evaluation of novel phthaloyl derivatives of 3-amino-3-aryl propionic acids as inhibitors of Trypanosoma cruzi trans-sialidase.Eur J Med Chem. 2018 Aug 5;156:252-268. doi: 10.1016/j.ejmech.2018.07.005. Epub 2018 Jul 7. Eur J Med Chem. 2018. PMID: 30006170
-
Trypanosoma cruzi trans-sialidase as a drug target against Chagas disease (American trypanosomiasis).Future Med Chem. 2013 Oct;5(15):1889-900. doi: 10.4155/fmc.13.129. Future Med Chem. 2013. PMID: 24144418 Review.
Cited by
-
Anti-Trypanosoma cruzi Properties of Sesquiterpene Lactones Isolated from Stevia spp.: In Vitro and In Silico Studies.Pharmaceutics. 2023 Feb 15;15(2):647. doi: 10.3390/pharmaceutics15020647. Pharmaceutics. 2023. PMID: 36839969 Free PMC article.
-
Insights from a comprehensive study of Trypanosoma cruzi: A new mitochondrial clade restricted to North and Central America and genetic structure of TcI in the region.PLoS Negl Trop Dis. 2021 Dec 17;15(12):e0010043. doi: 10.1371/journal.pntd.0010043. eCollection 2021 Dec. PLoS Negl Trop Dis. 2021. PMID: 34919556 Free PMC article.
References
-
- WHO. Chagas disease in Latin America: an epidemiological update based on 2010 estimates. 2015. - PubMed
-
- Zingales B, Miles MA, Campbell DA, Tibayrenc M, Macedo AM, Teixeira MMG, et al. The revised Trypanosoma cruzi subspecific nomenclature: Rationale, epidemiological relevance and research applications. Infection, Genetics and Evolution. 2012;12(2):240–53. http://dx.doi.org/10.1016/j.meegid.2011.12.009 - DOI - PubMed
-
- Moncayo A. Chagas disease: current epidemiological trends after the interruption of vectorial and transfusional transmission in the Southern Cone countries. Memórias do Instituto Oswaldo Cruz. 2003;98(5):577–91. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

