Posttranscriptional regulation of T helper cell fate decisions
- PMID: 29685903
- PMCID: PMC6080923
- DOI: 10.1083/jcb.201708075
Posttranscriptional regulation of T helper cell fate decisions
Abstract
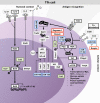
T helper cell subsets orchestrate context- and pathogen-specific responses of the immune system. They mostly do so by secreting specific cytokines that attract or induce activation and differentiation of other immune or nonimmune cells. The differentiation of T helper 1 (Th1), Th2, T follicular helper, Th17, and induced regulatory T cell subsets from naive T cells depends on the activation of intracellular signal transduction cascades. These cascades originate from T cell receptor and costimulatory receptor engagement and also receive critical input from cytokine receptors that sample the cytokine milieu within secondary lymphoid organs. Signal transduction then leads to the expression of subset-specifying transcription factors that, in concert with other transcription factors, up-regulate downstream signature genes. Although regulation of transcription is important, recent research has shown that posttranscriptional and posttranslational regulation can critically shape or even determine the outcome of Th cell differentiation. In this review, we describe how specific microRNAs, long noncoding RNAs, RNA-binding proteins, and ubiquitin-modifying enzymes regulate their targets to skew cell fate decisions.
© 2018 Hoefig and Heissmeyer.
Figures




Similar articles
-
Posttranscriptional Gene Regulation of T Follicular Helper Cells by RNA-Binding Proteins and microRNAs.Front Immunol. 2018 Jul 31;9:1794. doi: 10.3389/fimmu.2018.01794. eCollection 2018. Front Immunol. 2018. PMID: 30108596 Free PMC article. Review.
-
Rotenone Treatment Reveals a Role for Electron Transport Complex I in the Subcellular Localization of Key Transcriptional Regulators During T Helper Cell Differentiation.Front Immunol. 2018 Jun 7;9:1284. doi: 10.3389/fimmu.2018.01284. eCollection 2018. Front Immunol. 2018. PMID: 29930555 Free PMC article.
-
Regulation of human helper T cell subset differentiation by cytokines.Curr Opin Immunol. 2015 Jun;34:130-6. doi: 10.1016/j.coi.2015.03.007. Epub 2015 Apr 11. Curr Opin Immunol. 2015. PMID: 25879814 Free PMC article. Review.
-
Cytokine-modulated regulation of helper T cell populations.J Theor Biol. 2000 Oct 21;206(4):539-60. doi: 10.1006/jtbi.2000.2147. J Theor Biol. 2000. PMID: 11013114
-
Identification of global regulators of T-helper cell lineage specification.Genome Med. 2015 Nov 20;7:122. doi: 10.1186/s13073-015-0237-0. Genome Med. 2015. PMID: 26589177 Free PMC article.
Cited by
-
TTP-mediated regulation of mRNA stability in immune cells contributes to adaptive immunity, immune tolerance and clinical applications.RNA Biol. 2021 Dec;18(12):2150-2156. doi: 10.1080/15476286.2021.1917185. Epub 2021 Apr 27. RNA Biol. 2021. PMID: 33866923 Free PMC article. Review.
-
T cells at work: How post-transcriptional mechanisms control T cell homeostasis and activation.Eur J Immunol. 2021 Sep;51(9):2178-2187. doi: 10.1002/eji.202049055. Epub 2021 Jul 14. Eur J Immunol. 2021. PMID: 34180545 Free PMC article. Review.
-
Human T cells employ conserved AU-rich elements to fine-tune IFN-γ production.Eur J Immunol. 2020 Jul;50(7):949-958. doi: 10.1002/eji.201948458. Epub 2020 Mar 18. Eur J Immunol. 2020. PMID: 32112565 Free PMC article.
-
Anti-tumor and immune modulating activity of T cell induced tumor-targeting effectors (TITE).Cancer Immunol Immunother. 2021 Mar;70(3):633-656. doi: 10.1007/s00262-020-02692-8. Epub 2020 Aug 31. Cancer Immunol Immunother. 2021. PMID: 32865605 Free PMC article.
-
Post-transcriptional control of T-cell cytokine production: Implications for cancer therapy.Immunology. 2021 Sep;164(1):57-72. doi: 10.1111/imm.13339. Epub 2021 May 10. Immunology. 2021. PMID: 33884612 Free PMC article. Review.
References
-
- Annemann M., Wang Z., Plaza-Sirvent C., Glauben R., Schuster M., Ewald Sander F., Mamareli P., Kühl A.A., Siegmund B., Lochner M., and Schmitz I.. 2015. IκBNS regulates murine Th17 differentiation during gut inflammation and infection. J. Immunol. 194:2888–2898. 10.4049/jimmunol.1401964 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

