Domains within RbpA Serve Specific Functional Roles That Regulate the Expression of Distinct Mycobacterial Gene Subsets
- PMID: 29686140
- PMCID: PMC5996690
- DOI: 10.1128/JB.00690-17
Domains within RbpA Serve Specific Functional Roles That Regulate the Expression of Distinct Mycobacterial Gene Subsets
Abstract
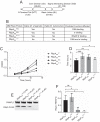
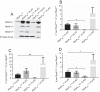
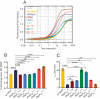
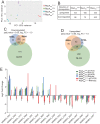
The RNA polymerase (RNAP) binding protein A (RbpA) contributes to the formation of stable RNAP-promoter open complexes (RPo) and is essential for viability in mycobacteria. Four domains have been identified in the RbpA protein, i.e., an N-terminal tail (NTT) that interacts with RNAP β' and σ subunits, a core domain (CD) that contacts the RNAP β' subunit, a basic linker (BL) that binds DNA, and a σ-interaction domain (SID) that binds group I and group II σ factors. Limited in vivo studies have been performed in mycobacteria, however, and how individual structural domains of RbpA contribute to RbpA function and mycobacterial gene expression remains mostly unknown. We investigated the roles of the RbpA structural domains in mycobacteria using a panel of rbpA mutants that target individual RbpA domains. The function of each RbpA domain was required for Mycobacterium tuberculosis viability and optimal growth in Mycobacterium smegmatis We determined that the RbpA SID is both necessary and sufficient for RbpA interaction with the RNAP, indicating that the primary functions of the NTT and CD are not solely association with the RNAP. We show that the RbpA BL and SID are required for RPo stabilization in vitro, while the NTT and CD antagonize this activity. Finally, RNA-sequencing analyses suggest that the NTT and CD broadly activate gene expression, whereas the BL and SID activate or repress gene expression in a gene-dependent manner for a subset of mycobacterial genes. Our findings highlight specific outcomes for the activities of the individual functional domains in RbpA.IMPORTANCEMycobacterium tuberculosis is the causative agent of tuberculosis and continues to be the most lethal infectious disease worldwide. Improved molecular understanding of the essential proteins involved in M. tuberculosis transcription, such as RbpA, could provide targets for much needed future therapeutic agents aimed at combatting this pathogen. In this study, we expand our understanding of RbpA by identifying the RbpA structural domains responsible for the interaction of RbpA with the RNAP and the effects of RbpA on transcription initiation and gene expression. These experiments expand our knowledge of RbpA while also broadening our understanding of bacterial transcription in general.
Keywords: Mycobacterium; RNA polymerases; RbpA; eubacteria; transcription; transcriptional regulation.
Copyright © 2018 American Society for Microbiology.
Figures
References
-
- World Health Organization. 2016. Global tuberculosis report 2016. World Health Organization, Geneva, Switzerland: http://apps.who.int/iris/bitstream/handle/10665/250441/9789241565394-eng....
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

