Community-driven roadmap for integrated disease maps
- PMID: 29688273
- PMCID: PMC6556900
- DOI: 10.1093/bib/bby024
Community-driven roadmap for integrated disease maps
Abstract
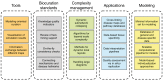
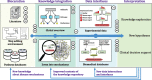
The Disease Maps Project builds on a network of scientific and clinical groups that exchange best practices, share information and develop systems biomedicine tools. The project aims for an integrated, highly curated and user-friendly platform for disease-related knowledge. The primary focus of disease maps is on interconnected signaling, metabolic and gene regulatory network pathways represented in standard formats. The involvement of domain experts ensures that the key disease hallmarks are covered and relevant, up-to-date knowledge is adequately represented. Expert-curated and computer readable, disease maps may serve as a compendium of knowledge, allow for data-supported hypothesis generation or serve as a scaffold for the generation of predictive mathematical models. This article summarizes the 2nd Disease Maps Community meeting, highlighting its important topics and outcomes. We outline milestones on the roadmap for the future development of disease maps, including creating and maintaining standardized disease maps; sharing parts of maps that encode common human disease mechanisms; providing technical solutions for complexity management of maps; and Web tools for in-depth exploration of such maps. A dedicated discussion was focused on mathematical modeling approaches, as one of the main goals of disease map development is the generation of mathematically interpretable representations to predict disease comorbidity or drug response and to suggest drug repositioning, altogether supporting clinical decisions.
Keywords: biocuration; disease maps; knowledge repository; mathematical modeling; molecular biology; pathway representation; translational medicine.
© The Author(s) 2018. Published by Oxford University Press.
Figures
References
-
- Niarakis A, Bounab Y, Grieco L, et al.Computational modeling of the main signaling pathways involved in mast cell activation. Curr Top Microbiol Immunol 2014;382:69–93. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

