Identification of a unique transcriptional architecture for the sigS operon in Staphylococcus aureus
- PMID: 29688345
- PMCID: PMC5974778
- DOI: 10.1093/femsle/fny108
Identification of a unique transcriptional architecture for the sigS operon in Staphylococcus aureus
Abstract
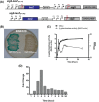
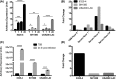
Staphylococcus aureus possess three alternative σ factors, including a lone extracytoplasmic function σ factor, σS. Our group previously identified and characterized this element, mapping three sigS promoters, demonstrating its inducibility during stress and virulence inducing conditions and demonstrating a role for this factor in disease causation. In the present study, we identify a fourth promoter of the sigS operon, termed P4, located in a unique position internal to the sigS coding region. Transcriptional profiling revealed that expression from P4 is dominant to the three upstream promoters, particularly upon exposure to chemical stressors that elicit DNA damage and disrupt cell wall stability; each of which have previously been shown to stimulate sigS expression. Importantly, expression of this fourth promoter, followed by at least one or more of the upstream promoters, is induced during growth in serum and upon phagocytosis by RAW 264.7 murine macrophage-like cells. Finally, we demonstrate that a downstream gene, SACOL1829, bears a large 3΄ UTR that spans the sigS-SACOL1828 coding region, and may serve to compete with the P4 transcript to inhibit σS production. Collectively, these findings reveal a unique operon architecture for the sigS locus that indicates the potential for novel regulatory mechanisms governing its expression.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

