Worldwide Protein Data Bank biocuration supporting open access to high-quality 3D structural biology data
- PMID: 29688351
- PMCID: PMC5804564
- DOI: 10.1093/database/bay002
Worldwide Protein Data Bank biocuration supporting open access to high-quality 3D structural biology data
Abstract
https://www.wwpdb.org/.
Figures


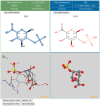

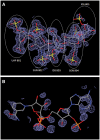
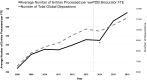
References
-
- Berman H.M., Henrick K., Nakamura H. (2003) Announcing the worldwide Protein Data Bank. Nat. Struct. Biol., 10, 980.. - PubMed
Publication types
MeSH terms
Grants and funding
- MR/L007835/1/MRC_/Medical Research Council/United Kingdom
- BB/M020428/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M011674/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- R01 GM109046/GM/NIGMS NIH HHS/United States
- BB/M013146/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 88944/WT_/Wellcome Trust/United Kingdom
- BB/G022577/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M020347/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 75968/WT_/Wellcome Trust/United Kingdom
- BB/K020013/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/K016970/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 104948/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

