A novel field-based molecular assay to detect validated artemisinin-resistant k13 mutants
- PMID: 29690890
- PMCID: PMC5916714
- DOI: 10.1186/s12936-018-2329-y
A novel field-based molecular assay to detect validated artemisinin-resistant k13 mutants
Abstract
Background: Given the risk of artemisinin resistance spreading from the Greater Mekong sub-region, prospective monitoring in sub-Saharan Africa should be expedited. Molecular biology techniques used for monitoring rely on the detection of k13 validated mutants by using PCR and Sanger sequencing approach, usually not available in malaria endemic areas.
Methods: A semi-automated workflow based on the easyMAG® platform and the Argene Solution® (bioMérieux, Marcy l'Etoile, France) as a field-based surveillance tool operable at national level was developed in four steps. Clinical and analytical performances of this tool detecting five of the most frequent and validated k13 mutants (Y493H, I543T, R539T, F446I and C580Y) from dried blood spots (DBS) were compared to the gold standard approach (PCR and Sanger sequencing).
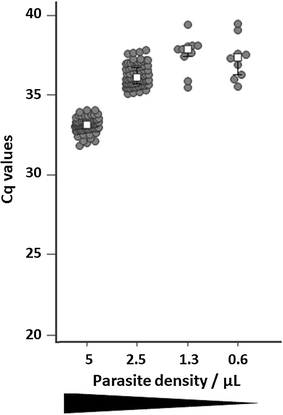
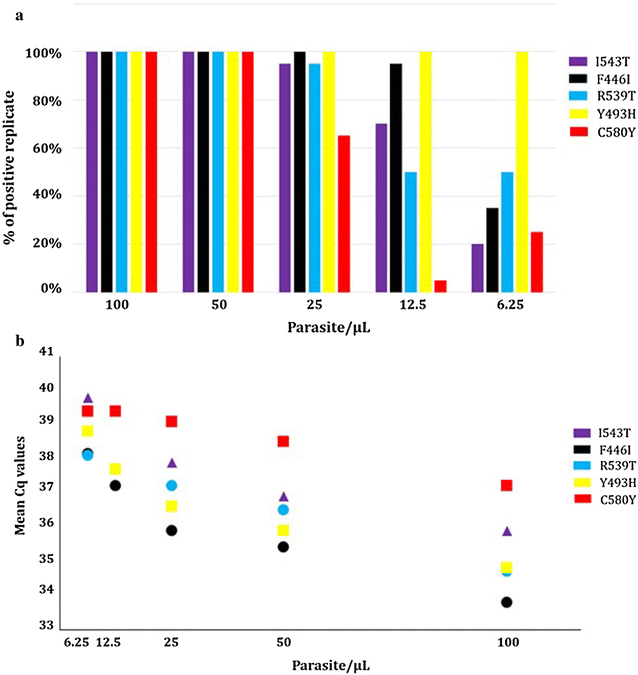
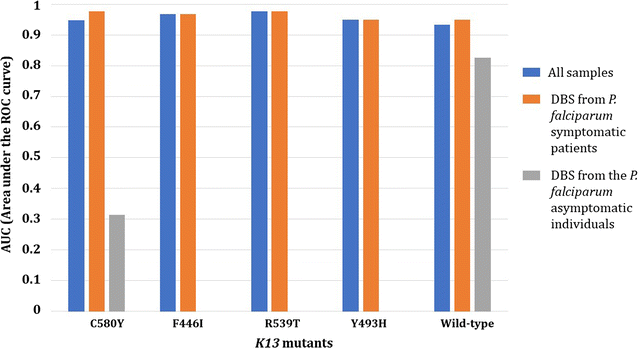
Results: By using the ARMS (amplification-refractory mutation system) strategy, the best multiplexing options were found in 3 separate real-time PCR duplexes (IC as internal control/I543T, C580Y/Y493H and F446I/R539T) with limits of detection ranging from 50 (C580Y) to 6.25 parasites/µL (Y493H). In field conditions, using 642 clinical DBS (from symptomatic patients and asymptomatic individuals) collected from Cambodia, Myanmar and Africa (Chad), the overall sensitivity and specificity of the K13 bMx prototype assay developed by bioMérieux were ≥ 90%. Areas under the ROC curves were estimated to be > 0.90 for all k13 mutants in samples from symptomatic patients.
Conclusion: The K13 ready-to-use bMx prototype assay, considered by the end-users as a user-friendly assay to perform (in shorter time than the K13 reference assay) and easy to interpret, was found to require less budget planning and had fewer logistical constraints. Its excellent performance qualifies the prototype as a reliable screening tool usable in malaria endemic countries recognized to be at risk of emergence or spread of validated k13 mutants. Additional multi-site studies are needed to evaluate the performances of the K13 bMx prototype assay in different epidemiological contexts such as Africa, India, or South America.
Keywords: Artemisinin resistance; Malaria; Plasmodium falciparum; Surveillance; k13 mutation detection.
Figures
Similar articles
-
Plasmodium berghei K13 Mutations Mediate In Vivo Artemisinin Resistance That Is Reversed by Proteasome Inhibition.mBio. 2020 Nov 10;11(6):e02312-20. doi: 10.1128/mBio.02312-20. mBio. 2020. PMID: 33173001 Free PMC article.
-
Prevalence of K13 mutation and Day-3 positive parasitaemia in artemisinin-resistant malaria endemic area of Cambodia: a cross-sectional study.Malar J. 2017 Sep 13;16(1):372. doi: 10.1186/s12936-017-2024-4. Malar J. 2017. PMID: 28903755 Free PMC article.
-
No evidence of amplified Plasmodium falciparum plasmepsin II gene copy number in an area with artemisinin-resistant malaria along the China-Myanmar border.Malar J. 2020 Sep 14;19(1):334. doi: 10.1186/s12936-020-03410-6. Malar J. 2020. PMID: 32928233 Free PMC article.
-
Importance of kelch 13 C580Y mutation in the studies of artemisinin resistance in Plasmodium falciparum in Greater Mekong Subregion.J Microbiol Immunol Infect. 2020 Oct;53(5):676-681. doi: 10.1016/j.jmii.2019.07.006. Epub 2019 Sep 10. J Microbiol Immunol Infect. 2020. PMID: 31563454 Review.
-
Molecular insights into artemisinin resistance in Plasmodium falciparum: An updated review.Infect Genet Evol. 2023 Aug;112:105460. doi: 10.1016/j.meegid.2023.105460. Epub 2023 Jun 1. Infect Genet Evol. 2023. PMID: 37269964 Review.
Cited by
-
A novel CRISPR-based malaria diagnostic capable of Plasmodium detection, species differentiation, and drug-resistance genotyping.EBioMedicine. 2021 Jun;68:103415. doi: 10.1016/j.ebiom.2021.103415. Epub 2021 Jun 14. EBioMedicine. 2021. PMID: 34139428 Free PMC article.
-
CRISPR-Based Programmable Nucleic Acid-Binding Protein Technology Can Specifically Detect Fatal Tropical Disease-Causing Pathogens.J Trop Med. 2022 Jul 18;2022:5390685. doi: 10.1155/2022/5390685. eCollection 2022. J Trop Med. 2022. PMID: 36199433 Free PMC article. Review.
-
Detection of Single-Nucleotide Polymorphism Markers of Antimalarial Drug Resistance Directly from Whole Blood.J Mol Diagn. 2019 Jul;21(4):623-631. doi: 10.1016/j.jmoldx.2019.02.004. Epub 2019 Jun 13. J Mol Diagn. 2019. PMID: 31204166 Free PMC article.
-
Evaluation of Artesunate-mefloquine as a Novel Alternative Treatment for Schistosomiasis in African Children (SchistoSAM): protocol of a proof-of-concept, open-label, two-arm, individually-randomised controlled trial.BMJ Open. 2021 Jun 24;11(6):e047147. doi: 10.1136/bmjopen-2020-047147. BMJ Open. 2021. PMID: 34168029 Free PMC article.
References
-
- WHO . World Malaria Report. Geneva: World Health Organization; 2017.
-
- Nosten F, White NJ. Artemisinin-based combination treatment of falciparum malaria. Am J Trop Med Hyg. 2007;77:181–192. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

