Heterologous DNA prime-protein boost immunization with RecA and FliD offers cross-clade protection against leptospiral infection
- PMID: 29691454
- PMCID: PMC5915591
- DOI: 10.1038/s41598-018-24674-8
Heterologous DNA prime-protein boost immunization with RecA and FliD offers cross-clade protection against leptospiral infection
Abstract
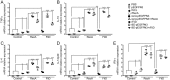
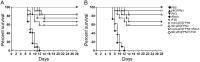
The emergence of >300 serovars of Leptospira confounded the use of generalized bacterin, the whole cell lysate, as vaccines to control leptospirosis. Because of substantial genetic and geographic heterogeneity among circulating serovars, one vaccine strain per serovar cannot be efficacious against all the serovars. We have performed heterologous DNA prime-protein boost vaccination challenge studies in hamsters using in vivo expressed, leptospiral recombinase A (RecA) and flagellar hook associated protein (FliD). We prepared the monovalent recombinant protein, plasmid DNA, and DNA prime protein boost adjuvant vaccines. The whole cell bacterin served as a control. Our data show that (i) RecA and FliD have multiple immunogenic B and T-cell epitopes with highly conserved domains among most prevalent pathogenic Leptospira spp., (ii) humoral and cell mediated immune responses were induced remarkably, (iii) provides significant protection against homologous (Autumnalis strain N2) and cross-clade heterologous (Canicola strain PAI-1) challenge infection for the heterologous prime-protein boost (∼91-100%) and, the DNA vaccine (∼75-83%). Recombinant protein vaccine shows only partial protection (∼58-66%), (iv) RecA prime-protein boost vaccine shows sterilizing immunity, with heterologous protection. This RecA/FliD prime-protein boost strategy holds potential for vaccination against animal leptospirosis and for a better control of zoonotic transmission.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Natarajaseenivasan K, et al. Seroprevalence of Leptospira borgpetersenii serovar javanica infection among dairy cattle, rats and humans in the Cauvery river valley of southern India. Southeast Asian J Trop Med Public Health. 2011;42:679–686. - PubMed
-
- WHO. Leptospirosis: an emerging public health problem. Wkly Epidemiol Rec/Heal Sect Secr Leag Nations45–52 (2011). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

