A traditional evolutionary history of foot-and-mouth disease viruses in Southeast Asia challenged by analyses of non-structural protein coding sequences
- PMID: 29691483
- PMCID: PMC5915611
- DOI: 10.1038/s41598-018-24870-6
A traditional evolutionary history of foot-and-mouth disease viruses in Southeast Asia challenged by analyses of non-structural protein coding sequences
Abstract
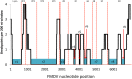
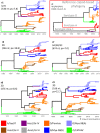
Recombination of rapidly evolving RNA-viruses provides an important mechanism for diversification, spread, and emergence of new variants with enhanced fitness. Foot-and-mouth disease virus (FMDV) causes an important transboundary disease of livestock that is endemic to most countries in Asia and Africa. Maintenance and spread of FMDV are driven by periods of dominance of specific viral lineages. Current understanding of the molecular epidemiology of FMDV lineages is generally based on the phylogenetic relationship of the capsid-encoding genes, with less attention to the process of recombination and evolution of non-structural proteins. In this study, the putative recombination breakpoints of FMDVs endemic to Southeast Asia were determined using full-open reading frame sequences. Subsequently, the lineages' divergence times of recombination-free genome regions were estimated. These analyses revealed a close relationship between two of the earliest endemic viral lineages that appear unrelated when only considering the phylogeny of their capsid proteins. Contrastingly, one lineage, named O/CATHAY, known for having a particular host predilection (pigs) has evolved independently. Additionally, intra-lineage recombination occurred at different breakpoints compared to the inter-lineage process. These results provide new insights about FMDV recombination patterns and the evolutionary interdependence of FMDV serotypes and lineages.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Reconstructing the evolutionary history of pandemic foot-and-mouth disease viruses: the impact of recombination within the emerging O/ME-SA/Ind-2001 lineage.Sci Rep. 2018 Oct 2;8(1):14693. doi: 10.1038/s41598-018-32693-8. Sci Rep. 2018. PMID: 30279570 Free PMC article.
-
Phylogeography of foot-and-mouth disease virus types O and A in Malaysia and surrounding countries.Infect Genet Evol. 2011 Mar;11(2):320-8. doi: 10.1016/j.meegid.2010.11.003. Epub 2010 Nov 18. Infect Genet Evol. 2011. PMID: 21093614
-
Comparative sequence analysis of representative foot-and-mouth disease virus genomes from Southeast Asia.Virus Genes. 2011 Aug;43(1):41-5. doi: 10.1007/s11262-011-0599-3. Epub 2011 Apr 9. Virus Genes. 2011. PMID: 21479678
-
Combining livestock trade patterns with phylogenetics to help understand the spread of foot and mouth disease in sub-Saharan Africa, the Middle East and Southeast Asia.Rev Sci Tech. 2011 Apr;30(1):63-85. doi: 10.20506/rst.30.1.2022. Rev Sci Tech. 2011. PMID: 21809754 Review.
-
Review of the Global Distribution of Foot-and-Mouth Disease Virus from 2007 to 2014.Transbound Emerg Dis. 2017 Apr;64(2):316-332. doi: 10.1111/tbed.12373. Epub 2015 May 20. Transbound Emerg Dis. 2017. PMID: 25996568 Review.
Cited by
-
Risk of transmission of foot-and-mouth disease by wild animals: infection dynamics in Japanese wild boar following direct inoculation or contact exposure.Vet Res. 2022 Oct 22;53(1):86. doi: 10.1186/s13567-022-01106-0. Vet Res. 2022. PMID: 36273214 Free PMC article.
-
Early origin and global colonisation of foot-and-mouth disease virus.Sci Rep. 2020 Sep 17;10(1):15268. doi: 10.1038/s41598-020-72246-6. Sci Rep. 2020. PMID: 32943727 Free PMC article.
-
Use of Synonymous Deoptimization to Derive Modified Live Attenuated Strains of Foot and Mouth Disease Virus.Front Microbiol. 2021 Jan 21;11:610286. doi: 10.3389/fmicb.2020.610286. eCollection 2020. Front Microbiol. 2021. PMID: 33552021 Free PMC article.
-
Foot-and-Mouth Disease Virus Interserotypic Recombination in Superinfected Carrier Cattle.Pathogens. 2022 Jun 3;11(6):644. doi: 10.3390/pathogens11060644. Pathogens. 2022. PMID: 35745498 Free PMC article.
-
Preparation and Characterization of Nanoliposome Containing Isolated VP1 Protein of Foot and Mouth Disease Virus as a Model of Vaccine.Arch Razi Inst. 2022 Feb 28;77(1):37-44. doi: 10.22092/ari.2021.353322.1596. eCollection 2022 Feb. Arch Razi Inst. 2022. PMID: 35891774 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

