Lactobacilli Are Prominent Members of the Microbiota Involved in the Ruminal Digestion of Barley and Corn
- PMID: 29692773
- PMCID: PMC5902705
- DOI: 10.3389/fmicb.2018.00718
Lactobacilli Are Prominent Members of the Microbiota Involved in the Ruminal Digestion of Barley and Corn
Abstract
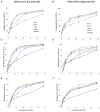
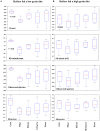
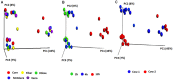
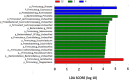
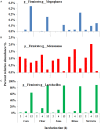
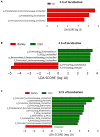
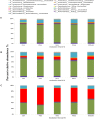
The chemical composition of barley grain can vary among barley varieties (Fibar, Xena, McGwire, and Hilose) and result in different digestion efficiencies in the rumen. It is not known if compositional differences in barley can affect the microbiota involved in the ruminal digestion of barley. The objective of this study was to characterize the in situ rumen degradability and microbiota of four barley grain varieties and to compare these to corn. Three ruminally cannulated heifers were fed a low (60% barley silage, 37% barley grain, and 3% supplement) or high grain (37% barley silage, 60% barley grain, and 3% supplement) diet. One set of bags was used to estimate dry matter (DM), starch and crude protein (CP) degradability. A second set was used to extract DNA from the adherent microbiota and visualize grain after incubation using scanning electron microscopy (SEM). DNA was subjected to amplicon 16S rRNA gene sequencing followed by analysis using QIIME. In the low grain diet, McGwire had the highest effective degradability (ED) of DM (P < 0.01). The ED of starch was highest (P < 0.01) for Fibar, McGwire, and Xena, but the ED of CP was not affected by variety. For the high grain diet, Xena and McGwire had the highest ED of DM (P < 0.01). The ED of starch was highest (P < 0.01) for Xena and Fibar. The ED of protein was highest (P < 0.01) for Xena and McGwire. Although the microbiota did not differ among barley varieties, they did differ from corn and with incubation time. Lactobacilli were dominant members of the mature biofilms associated with corn and barley and were accompanied by a notable increase in the lactic acid utilizing genera, Megasphaera. As none of the cattle exhibited subclinical or clinical acidosis during the study, our results suggest that lactobacilli play a more prominent role in routine starch digestion than presently surmised.
Keywords: barley; biofilm; cattle; corn; microbiome; rumen; starch digestion.
Figures


References
-
- Åman P., Newman C. W. (1986). Chemical composition of some different types of barley grown in Montana, USA. J. Cereal Sci. 4 133–141. 10.1016/S0733-5210(86)80016-9 - DOI
-
- AOAC (1997). Official Methods of Analysis of the Association of Official Analytical Chemists 20th Edn. Washington DC: Association of Official Analytical Chemists.
-
- AOAC (2010). Official Methods of Analysis of Official Analytical Chemists 18th Edn. Arlington, VA: Association of Official Analytical Chemists.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

