Genome-Wide Association Analyses Highlight the Potential for Different Genetic Mechanisms for Litter Size Among Sheep Breeds
- PMID: 29692799
- PMCID: PMC5902979
- DOI: 10.3389/fgene.2018.00118
Genome-Wide Association Analyses Highlight the Potential for Different Genetic Mechanisms for Litter Size Among Sheep Breeds
Abstract
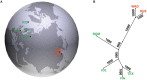
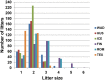
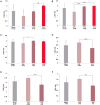
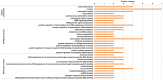
Reproduction is an important trait in sheep breeding as well as in other livestock. However, despite its importance the genetic mechanisms of litter size in domestic sheep (Ovis aries) are still poorly understood. To explore genetic mechanisms underlying the variation in litter size, we conducted multiple independent genome-wide association studies in five sheep breeds of high prolificacy (Wadi, Hu, Icelandic, Finnsheep, and Romanov) and one low prolificacy (Texel) using the Ovine Infinium HD BeadChip, respectively. We identified different sets of candidate genes associated with litter size in different breeds: BMPR1B, FBN1, and MMP2 in Wadi; GRIA2, SMAD1, and CTNNB1 in Hu; NCOA1 in Icelandic; INHBB, NF1, FLT1, PTGS2, and PLCB3 in Finnsheep; ESR2 in Romanov and ESR1, GHR, ETS1, MMP15, FLI1, and SPP1 in Texel. Further annotation of genes and bioinformatics analyses revealed that different biological pathways could be involved in the variation in litter size of females: hormone secretion (FSH and LH) in Wadi and Hu, placenta and embryonic lethality in Icelandic, folliculogenesis and LH signaling in Finnsheep, ovulation and preovulatory follicle maturation in Romanov, and estrogen and follicular growth in Texel. Taken together, our results provide new insights into the genetic mechanisms underlying the prolificacy trait in sheep and other mammals, suggesting targets for selection where the aim is to increase prolificacy in breeding projects.
Keywords: biological pathways; genome-wide association study; prolificacy; regulation; sheep.
Figures





References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

