A New Approach to Assessing HSV-1 Recombination during Intercellular Spread
- PMID: 29693602
- PMCID: PMC5977213
- DOI: 10.3390/v10050220
A New Approach to Assessing HSV-1 Recombination during Intercellular Spread
Abstract
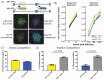
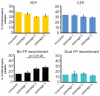
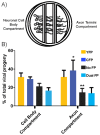
The neuroinvasive Herpes simplex virus type 1 (HSV-1) utilizes intergenomic recombination in order to diversify viral populations. Research efforts to assess HSV-1 recombination are often complicated by the use of attenuating mutations, which differentiate viral progeny but unduly influence the replication and spread. In this work, we generated viruses with markers that allowed for classification of viral progeny with limited attenuation of viral replication. We isolated viruses, harboring either a cyan (C) or yellow (Y) fluorescent protein (FP) expression cassette inserted in two different locations within the viral genome, in order to visually quantify the recombinant progeny based on plaque fluorescence. We found that the FP marked genomes had a limited negative affect on the viral replication and production of progeny virions. A co-infection of the two viruses resulted in recombinant progeny that was dependent on the multiplicity of infection and independent of the time post infection, at a rate that was similar to previous reports. The sequential passage of mixed viral populations revealed a limited change in the distribution of the parental and recombinant progeny. Interestingly, the neuroinvasive spread within neuronal cultures and an in vivo mouse model, revealed large, random shifts in the parental and recombinant distributions in viral populations. In conclusion, our approach highlights the utility of FP expressing viruses in order to provide new insights into mechanisms of HSV-1 recombination.
Keywords: HSV-1; alphaherpesvirus; cell–cell spread; fluorescent protein; intravitreal injection; neuroinvasion; neuron culture; recombination.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
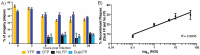

Similar articles
-
A dual fluorescent herpes simplex virus type 1 recombinant reveals divergent outcomes of neuronal infection.J Virol. 2024 May 14;98(5):e0003224. doi: 10.1128/jvi.00032-24. Epub 2024 Apr 23. J Virol. 2024. PMID: 38651900 Free PMC article.
-
Herpes Simplex Virus 1 Coinfection Modifies Adeno-associated Virus Genome End Recombination.J Virol. 2021 Jun 10;95(13):e0048621. doi: 10.1128/JVI.00486-21. Epub 2021 Jun 10. J Virol. 2021. PMID: 33853961 Free PMC article.
-
gD-Independent Superinfection Exclusion of Alphaherpesviruses.J Virol. 2016 Mar 28;90(8):4049-58. doi: 10.1128/JVI.00089-16. Print 2016 Apr. J Virol. 2016. PMID: 26842480 Free PMC article.
-
Deletion of the Herpes simplex 1 internal repeat sequences affects pathogenicity in the mouse.Front Biosci. 1996 Oct 4;1:a59-68. doi: 10.2741/a106. Front Biosci. 1996. PMID: 9159195
-
Re-evaluating natural resistance to herpes simplex virus type 1.J Virol. 2004 Sep;78(18):10086-95. doi: 10.1128/JVI.78.18.10086-10095.2004. J Virol. 2004. PMID: 15331741 Free PMC article.
Cited by
-
Viral Recombination: Ecology, Evolution, and Pathogenesis.Viruses. 2018 Jul 6;10(7):358. doi: 10.3390/v10070358. Viruses. 2018. PMID: 29986376 Free PMC article.
-
A Single Herpes Simplex Virus 1 Genome Reactivates from Individual Cells.Microbiol Spectr. 2022 Aug 31;10(4):e0114422. doi: 10.1128/spectrum.01144-22. Epub 2022 Jul 11. Microbiol Spectr. 2022. PMID: 35862979 Free PMC article.
-
A dual fluorescent herpes simplex virus type 1 recombinant reveals divergent outcomes of neuronal infection.J Virol. 2024 May 14;98(5):e0003224. doi: 10.1128/jvi.00032-24. Epub 2024 Apr 23. J Virol. 2024. PMID: 38651900 Free PMC article.
-
Viral gene drive spread during herpes simplex virus 1 infection in mice.Nat Commun. 2024 Sep 17;15(1):8161. doi: 10.1038/s41467-024-52395-2. Nat Commun. 2024. PMID: 39289368 Free PMC article.
-
Herpes simplex virus 1 strain 17+ with R2 mutation in UL37 has residual retrograde transport.bioRxiv [Preprint]. 2025 Jun 26:2025.06.25.661543. doi: 10.1101/2025.06.25.661543. bioRxiv. 2025. PMID: 40667132 Free PMC article. Preprint.
References
-
- Loncoman C.A., Vaz P.K., Coppo M.J., Hartley C.A., Morera F.J., Browning G.F., Devlin J.M. Natural recombination in alphaherpesviruses: Insights into viral evolution through full genome sequencing and sequence analysis. Infect. Genet. Evol. 2017;49:174–185. doi: 10.1016/j.meegid.2016.12.022. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

