Exploration of gene functions for esophageal squamous cell carcinoma using network-based guilt by association principle
- PMID: 29694510
- PMCID: PMC5937724
- DOI: 10.1590/1414-431x20186801
Exploration of gene functions for esophageal squamous cell carcinoma using network-based guilt by association principle
Abstract
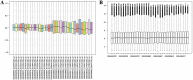
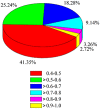
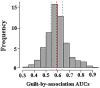
Gene networks have been broadly used to predict gene functions based on guilt by association (GBA) principle. Thus, in order to better understand the molecular mechanisms of esophageal squamous cell carcinoma (ESCC), our study was designed to use a network-based GBA method to identify the optimal gene functions for ESCC. To identify genomic bio-signatures for ESCC, microarray data of GSE20347 were first downloaded from a public functional genomics data repository of Gene Expression Omnibus database. Then, differentially expressed genes (DEGs) between ESCC patients and controls were identified using the LIMMA method. Afterwards, construction of differential co-expression network (DCN) was performed relying on DEGs, followed by gene ontology (GO) enrichment analysis based on a known confirmed database and DEGs. Eventually, the optimal gene functions were predicted using GBA algorithm based on the area under the curve (AUC) for each GO term. Overall, 43 DEGs and 67 GO terms were gained for subsequent analysis. GBA predictions demonstrated that 13 GO functions with AUC>0.7 had a good classification ability. Significantly, 6 out of 13 GO terms yielded AUC>0.8, which were determined as the optimal gene functions. Interestingly, there were two GO categories with AUC>0.9, which included cell cycle checkpoint (AUC=0.91648), and mitotic sister chromatid segregation (AUC=0.91597). Our findings highlight the clinical implications of cell cycle checkpoint and mitotic sister chromatid segregation in ESCC progression and provide the molecular foundation for developing therapeutic targets.
Figures

References
-
- Wong GS. Periostin and its role in promoting invasion in the tumor microenvironment of esophageal cancer. University of Pennsylvania, ProQuest Dissertations Publishing; 2012. p. 3551813.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

