High-Dimensional Phenotyping Identifies Age-Emergent Cells in Human Mammary Epithelia
- PMID: 29694896
- PMCID: PMC5946804
- DOI: 10.1016/j.celrep.2018.03.114
High-Dimensional Phenotyping Identifies Age-Emergent Cells in Human Mammary Epithelia
Abstract
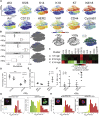
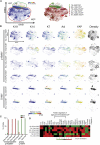
Aging is associated with tissue-level changes in cellular composition that are correlated with increased susceptibility to disease. Aging human mammary tissue shows skewed progenitor cell potency, resulting in diminished tumor-suppressive cell types and the accumulation of defective epithelial progenitors. Quantitative characterization of these age-emergent human cell subpopulations is lacking, impeding our understanding of the relationship between age and cancer susceptibility. We conducted single-cell resolution proteomic phenotyping of healthy breast epithelia from 57 women, aged 16-91 years, using mass cytometry. Remarkable heterogeneity was quantified within the two mammary epithelial lineages. Population partitioning identified a subset of aberrant basal-like luminal cells that accumulate with age and originate from age-altered progenitors. Quantification of age-emergent phenotypes enabled robust classification of breast tissues by age in healthy women. This high-resolution mapping highlighted specific epithelial subpopulations that change with age in a manner consistent with increased susceptibility to breast cancer.
Keywords: aging; breast cancer; heterogeneity; human mammary epithelia; mass cytometry; single-cell analysis.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Figures






References
-
- Bandura D.R., Baranov V.I., Ornatsky O.I., Antonov A., Kinach R., Lou X., Pavlov S., Vorobiev S., Dick J.E., Tanner S.D. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal. Chem. 2009;81:6813–6822. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

