CRISPR/Cas9-mediated genome editing in a reef-building coral
- PMID: 29695630
- PMCID: PMC5960312
- DOI: 10.1073/pnas.1722151115
CRISPR/Cas9-mediated genome editing in a reef-building coral
Abstract
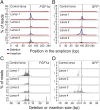
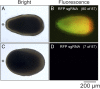
Reef-building corals are critically important species that are threatened by anthropogenic stresses including climate change. In attempts to understand corals' responses to stress and other aspects of their biology, numerous genomic and transcriptomic studies have been performed, generating a variety of hypotheses about the roles of particular genes and molecular pathways. However, it has not generally been possible to test these hypotheses rigorously because of the lack of genetic tools for corals. Here, we demonstrate efficient genome editing using the CRISPR/Cas9 system in the coral Acropora millepora We targeted the genes encoding fibroblast growth factor 1a (FGF1a), green fluorescent protein (GFP), and red fluorescent protein (RFP). After microinjecting CRISPR/Cas9 ribonucleoprotein complexes into fertilized eggs, we detected induced mutations in the targeted genes using changes in restriction-fragment length, Sanger sequencing, and high-throughput Illumina sequencing. We observed mutations in ∼50% of individuals screened, and the proportions of wild-type and various mutant gene copies in these individuals indicated that mutation induction continued for at least several cell cycles after injection. Although multiple paralogous genes encoding green fluorescent proteins are present in A. millepora, appropriate design of the guide RNA allowed us to induce mutations simultaneously in more than one paralog. Because A. millepora larvae can be induced to settle and begin colony formation in the laboratory, CRISPR/Cas9-based gene editing should allow rigorous tests of gene function in both larval and adult corals.
Keywords: Acropora millepora; CRISPR/Cas9; coral; genome editing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Moberg F, Folke C. Ecological goods and services of coral reef ecosystems. Ecol Econ. 1999;29:215–233.
-
- Hughes TP, et al. Coral reefs in the Anthropocene. Nature. 2017;546:82–90. - PubMed
-
- Muscatine L, Cernichiari E. Assimilation of photosynthetic products of zooxanthellae by a reef coral. Biol Bull. 1969;137:506–523. - PubMed
-
- Muscatine L, Falkowski PG, Porter JW, Dubinsky Z. Fate of photosynthetic fixed carbon in light- and shade-adapted colonies of the symbiotic coral Stylophora pistillata. Proc R Soc B. 1984;222:181–202.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

