Disease-causing variants in TCF4 are a frequent cause of intellectual disability: lessons from large-scale sequencing approaches in diagnosis
- PMID: 29695756
- PMCID: PMC6018712
- DOI: 10.1038/s41431-018-0096-4
Disease-causing variants in TCF4 are a frequent cause of intellectual disability: lessons from large-scale sequencing approaches in diagnosis
Abstract
High-throughput sequencing (HTS) of human genome coding regions allows the simultaneous screen of a large number of genes, significantly improving the diagnosis of non-syndromic intellectual disabilities (ID). HTS studies permit the redefinition of the phenotypical spectrum of known disease-causing genes, escaping the clinical inclusion bias of gene-by-gene Sanger sequencing. We studied a cohort of 903 patients with ID not reminiscent of a well-known syndrome, using an ID-targeted HTS of several hundred genes and found de novo heterozygous variants in TCF4 (transcription factor 4) in eight novel patients. Piecing together the patients from this study and those from previous large-scale unbiased HTS studies, we estimated the rate of individuals with ID carrying a disease-causing TCF4 mutation to 0.7%. So far, TCF4 molecular abnormalities were known to cause a syndromic form of ID, Pitt-Hopkins syndrome (PTHS), which combines severe ID, developmental delay, absence of speech, behavioral and ventilation disorders, and a distinctive facial gestalt. Therefore, we reevaluated ten patients carrying a pathogenic or likely pathogenic variant in TCF4 (eight patients included in this study and two from our previous ID-HTS study) for PTHS criteria defined by Whalen and Marangi. A posteriori, five patients had a score highly evocative of PTHS, three were possibly consistent with this diagnosis, and two had a score below the defined PTHS threshold. In conclusion, these results highlight TCF4 as a frequent cause of moderate to profound ID and broaden the clinical spectrum associated to TCF4 mutations to nonspecific ID.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
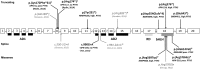

References
-
- Amiel J, Rio M, de Pontual L, et al. Mutations in TCF4, encoding a class I basic helix-loop-helix transcription factor, are responsible for Pitt-Hopkins syndrome, a severe epileptic encephalopathy associated with autonomic dysfunction. Am J Hum Genet. 2007;80:988–93. doi: 10.1086/515582. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources

