TFAP2C regulates transcription in human naive pluripotency by opening enhancers
- PMID: 29695788
- PMCID: PMC5926822
- DOI: 10.1038/s41556-018-0089-0
TFAP2C regulates transcription in human naive pluripotency by opening enhancers
Abstract
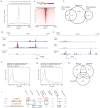
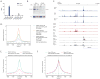
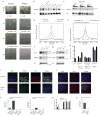
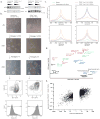
Naive and primed pluripotent human embryonic stem cells bear transcriptional similarity to pre- and post-implantation epiblast and thus constitute a developmental model for understanding the pluripotent stages in human embryo development. To identify new transcription factors that differentially regulate the unique pluripotent stages, we mapped open chromatin using ATAC-seq and found enrichment of the activator protein-2 (AP2) transcription factor binding motif at naive-specific open chromatin. We determined that the AP2 family member TFAP2C is upregulated during primed to naive reversion and becomes widespread at naive-specific enhancers. TFAP2C functions to maintain pluripotency and repress neuroectodermal differentiation during the transition from primed to naive by facilitating the opening of enhancers proximal to pluripotency factors. Additionally, we identify a previously undiscovered naive-specific POU5F1 (OCT4) enhancer enriched for TFAP2C binding. Taken together, TFAP2C establishes and maintains naive human pluripotency and regulates OCT4 expression by mechanisms that are distinct from mouse.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

