Uncovering inherent cellular plasticity of multiciliated ependyma leading to ventricular wall transformation and hydrocephalus
- PMID: 29695808
- PMCID: PMC5916891
- DOI: 10.1038/s41467-018-03812-w
Uncovering inherent cellular plasticity of multiciliated ependyma leading to ventricular wall transformation and hydrocephalus
Abstract
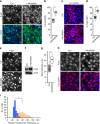
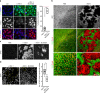
Specialized, differentiated cells often perform unique tasks that require them to maintain a stable phenotype. Multiciliated ependymal cells (ECs) are unique glial cells lining the brain ventricles, important for cerebral spinal fluid circulation. While functional ECs are needed to prevent hydrocephalus, they have also been reported to generate new neurons: whether ECs represent a stable cellular population remains unclear. Via a chemical screen we found that mature ECs are inherently plastic, with their multiciliated state needing constant maintenance by the Foxj1 transcription factor, which paradoxically is rapidly turned over by the ubiquitin-proteasome system leading to cellular de-differentiation. Mechanistic analyses revealed a novel NF-κB-independent IKK2 activity stabilizing Foxj1 in mature ECs, and we found that known IKK2 inhibitors including viruses and growth factors robustly induced Foxj1 degradation, EC de-differentiation, and hydrocephalus. Although mature ECs upon de-differentiation can divide and regenerate multiciliated ECs, we did not detect evidence supporting EC's neurogenic potential.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

