BitterSweetForest: A Random Forest Based Binary Classifier to Predict Bitterness and Sweetness of Chemical Compounds
- PMID: 29696137
- PMCID: PMC5905275
- DOI: 10.3389/fchem.2018.00093
BitterSweetForest: A Random Forest Based Binary Classifier to Predict Bitterness and Sweetness of Chemical Compounds
Abstract
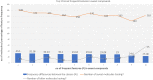
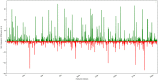
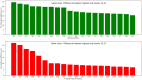
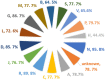
Taste of a chemical compound present in food stimulates us to take in nutrients and avoid poisons. However, the perception of taste greatly depends on the genetic as well as evolutionary perspectives. The aim of this work was the development and validation of a machine learning model based on molecular fingerprints to discriminate between sweet and bitter taste of molecules. BitterSweetForest is the first open access model based on KNIME workflow that provides platform for prediction of bitter and sweet taste of chemical compounds using molecular fingerprints and Random Forest based classifier. The constructed model yielded an accuracy of 95% and an AUC of 0.98 in cross-validation. In independent test set, BitterSweetForest achieved an accuracy of 96% and an AUC of 0.98 for bitter and sweet taste prediction. The constructed model was further applied to predict the bitter and sweet taste of natural compounds, approved drugs as well as on an acute toxicity compound data set. BitterSweetForest suggests 70% of the natural product space, as bitter and 10% of the natural product space as sweet with confidence score of 0.60 and above. 77% of the approved drug set was predicted as bitter and 2% as sweet with a confidence score of 0.75 and above. Similarly, 75% of the total compounds from acute oral toxicity class were predicted only as bitter with a minimum confidence score of 0.75, revealing toxic compounds are mostly bitter. Furthermore, we applied a Bayesian based feature analysis method to discriminate the most occurring chemical features between sweet and bitter compounds using the feature space of a circular fingerprint.
Keywords: KNIME workflow; Random Forest; bitter prediction; fingerprints; sweetness prediction; taste prediction.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources

