Trypanosomal mitochondrial intermediate peptidase does not behave as a classical mitochondrial processing peptidase
- PMID: 29698456
- PMCID: PMC5919513
- DOI: 10.1371/journal.pone.0196474
Trypanosomal mitochondrial intermediate peptidase does not behave as a classical mitochondrial processing peptidase
Abstract
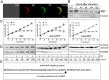
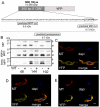
Upon their translocation into the mitochondrial matrix, the N-terminal pre-sequence of nuclear-encoded proteins undergoes cleavage by mitochondrial processing peptidases. Some proteins require more than a single processing step, which involves several peptidases. Down-regulation of the putative Trypanosoma brucei mitochondrial intermediate peptidase (MIP) homolog by RNAi renders the cells unable to grow after 48 hours of induction. Ablation of MIP results in the accumulation of the precursor of the trypanosomatid-specific trCOIV protein, the largest nuclear-encoded subunit of the cytochrome c oxidase complex in this flagellate. However, the trCOIV precursor of the same size accumulates also in trypanosomes in which either alpha or beta subunits of the mitochondrial processing peptidase (MPP) have been depleted. Using a chimeric protein that consists of the N-terminal sequence of a putative subunit of respiratory complex I fused to a yellow fluorescent protein, we assessed the accumulation of the precursor protein in trypanosomes, in which RNAi was induced against the alpha or beta subunits of MPP or MIP. The observed accumulation of precursors indicates MIP depletion affects the activity of the cannonical MPP, or at least one of its subunits.
Conflict of interest statement
Figures

Similar articles
-
Trypanosoma brucei has a canonical mitochondrial processing peptidase.Mol Biochem Parasitol. 2012 Oct;185(2):161-4. doi: 10.1016/j.molbiopara.2012.07.005. Epub 2012 Jul 25. Mol Biochem Parasitol. 2012. PMID: 22841752
-
An Advanced System of the Mitochondrial Processing Peptidase and Core Protein Family in Trypanosoma brucei and Multiple Origins of the Core I Subunit in Eukaryotes.Genome Biol Evol. 2013;5(5):860-75. doi: 10.1093/gbe/evt056. Genome Biol Evol. 2013. PMID: 23563972 Free PMC article.
-
Mitochondrial processing peptidases.Biochim Biophys Acta. 2002 Sep 2;1592(1):63-77. doi: 10.1016/s0167-4889(02)00265-3. Biochim Biophys Acta. 2002. PMID: 12191769 Review.
-
Disparate phenotypic effects from the knockdown of various Trypanosoma brucei cytochrome c oxidase subunits.Mol Biochem Parasitol. 2012 Aug;184(2):90-8. doi: 10.1016/j.molbiopara.2012.04.013. Epub 2012 May 5. Mol Biochem Parasitol. 2012. PMID: 22569586
-
Mitochondrial processing peptidase: multiple-site recognition of precursor proteins.Biochem Biophys Res Commun. 1999 Nov 30;265(3):611-6. doi: 10.1006/bbrc.1999.1703. Biochem Biophys Res Commun. 1999. PMID: 10600469 Review.
Cited by
-
Expanded Proteomic Survey of the Human Parasite Leishmania major Focusing on Changes in Null Mutants of the Golgi GDP-Mannose/Fucose/Arabinopyranose Transporter LPG2 and of the Mitochondrial Fucosyltransferase FUT1.Microbiol Spectr. 2022 Dec 21;10(6):e0305222. doi: 10.1128/spectrum.03052-22. Epub 2022 Nov 17. Microbiol Spectr. 2022. PMID: 36394313 Free PMC article.
-
A comparison of three approaches for the discovery of novel tripartite attachment complex proteins in Trypanosoma brucei.PLoS Negl Trop Dis. 2020 Sep 16;14(9):e0008568. doi: 10.1371/journal.pntd.0008568. eCollection 2020 Sep. PLoS Negl Trop Dis. 2020. PMID: 32936798 Free PMC article.
-
An essential thioredoxin-type protein of Trypanosoma brucei acts as redox-regulated mitochondrial chaperone.PLoS Pathog. 2019 Sep 26;15(9):e1008065. doi: 10.1371/journal.ppat.1008065. eCollection 2019 Sep. PLoS Pathog. 2019. PMID: 31557263 Free PMC article.
-
The Remarkable Metabolism of Vickermania ingenoplastis: Genomic Predictions.Pathogens. 2021 Jan 14;10(1):68. doi: 10.3390/pathogens10010068. Pathogens. 2021. PMID: 33466586 Free PMC article.
-
Mechanisms of Insecticidal Action of Metarhizium anisopliae on Adult Japanese Pine Sawyer Beetles (Monochamus alternatus).ACS Omega. 2020 Sep 24;5(39):25312-25318. doi: 10.1021/acsomega.0c03585. eCollection 2020 Oct 6. ACS Omega. 2020. PMID: 33043210 Free PMC article.
References
-
- Neupert W. Protein import into mitochondria. Annu Rev Biochem. 1997;66: 863–917. doi: 10.1146/annurev.biochem.66.1.863 - DOI - PubMed
-
- Schmidt O, Pfanner N, Meisinger C. Mitochondrial protein import: from proteomics to functional mechanisms. Nat Rev Mol Cell Biol. 2010;11: 655–667. doi: 10.1038/nrm2959 - DOI - PubMed
-
- Hildenbeutel M, Habib SJ, Herrmann JM, Rapaport D. New insights into the mechanism of precursor protein insertion into the mitochondrial membranes. Int Rev Cell Mol Biol. 2008;268: 147–190. doi: 10.1016/S1937-6448(08)00805-8 - DOI - PubMed
-
- Eckers E, Cyrklaff M, Simpson L, Deponte M. Mitochondrial protein import pathways are functionally conserved among eukaryotes despite compositional diversity of the import machineries. Biol Chem. 2012;393: 513–524. doi: 10.1515/hsz-2011-0255 - DOI - PubMed
-
- Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138: 628–644. doi: 10.1016/j.cell.2009.08.005 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

