Megakaryocyte lineage development is controlled by modulation of protein acetylation
- PMID: 29698469
- PMCID: PMC5919413
- DOI: 10.1371/journal.pone.0196400
Megakaryocyte lineage development is controlled by modulation of protein acetylation
Abstract
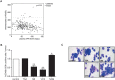
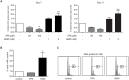
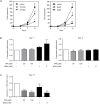
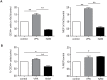
Treatment with lysine deacetylase inhibitors (KDACi) for haematological malignancies, is accompanied by haematological side effects including thrombocytopenia, suggesting that modulation of protein acetylation affects normal myeloid development, and specifically megakaryocyte development. In the current study, utilising ex-vivo differentiation of human CD34+ haematopoietic progenitor cells, we investigated the effects of two functionally distinct KDACi, valproic acid (VPA), and nicotinamide (NAM), on megakaryocyte differentiation, and lineage choice decisions. Treatment with VPA increased the number of megakaryocyte/erythroid progenitors (MEP), accompanied by inhibition of megakaryocyte differentiation, whereas treatment with NAM accelerated megakaryocyte development, and stimulated polyploidisation. Treatment with both KDACi resulted in no significant effects on erythrocyte differentiation, suggesting that the effects of KDACi primarily affect megakaryocyte lineage development. H3K27Ac ChIP-sequencing analysis revealed that genes involved in myeloid development, as well as megakaryocyte/erythroid (ME)-lineage differentiation are uniquely modulated by specific KDACi treatment. Taken together, our data reveal distinct effects of specific KDACi on megakaryocyte development, and ME-lineage decisions, which can be partially explained by direct effects on promoter acetylation of genes involved in myeloid differentiation.
Conflict of interest statement
Figures
References
-
- Rhodes J, Hagen A, Hsu K, Deng M, Liu TX, Look AT, et al. Interplay of pu.1 and gata1 determines myelo-erythroid progenitor cell fate in zebrafish. Developmental cell. 2005;8(1):97–108. doi: 10.1016/j.devcel.2004.11.014 - DOI - PubMed
-
- Levin J, Peng JP, Baker GR, Villeval JL, Lecine P, Burstein SA, et al. Pathophysiology of thrombocytopenia and anemia in mice lacking transcription factor NF-E2. Blood. 1999;94(9):3037–47. - PubMed
-
- Tijssen MR, Cvejic A, Joshi A, Hannah RL, Ferreira R, Forrai A, et al. Genome-wide analysis of simultaneous GATA1/2, RUNX1, FLI1, and SCL binding in megakaryocytes identifies hematopoietic regulators. Dev Cell 2011;20(5): 597–609. doi: 10.1016/j.devcel.2011.04.008 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

