Massively parallel sequencing of micro-manipulated cells targeting a comprehensive panel of disease-causing genes: A comparative evaluation of upstream whole-genome amplification methods
- PMID: 29698522
- PMCID: PMC5919401
- DOI: 10.1371/journal.pone.0196334
Massively parallel sequencing of micro-manipulated cells targeting a comprehensive panel of disease-causing genes: A comparative evaluation of upstream whole-genome amplification methods
Abstract
Single Gene Disorders (SGD) are still routinely diagnosed using PCR-based assays that need to be developed and validated for each individual disease-specific gene fragment. The TruSight One sequencing panel currently covers 12 Mb of genomic content, including 4813 genes associated with a clinical phenotype. When only a limited number of cells are available, whole genome amplification (WGA) is required prior to DNA target capture techniques such as the TruSight One panel. In this study, we compared 4 different WGA methods in combination with the TruSight One sequencing panel to perform single nucleotide polymorphism (SNP) genotyping starting from 3 micro-manipulated cells. This setting simulates clinical settings such as day-5 blastocyst biopsy for Preimplantation Genetic Testing (PGT), liquid biopsy of circulating tumor cells (CTCs) and cancer-cell profiling. Bulk cell samples were processed alongside these WGA samples to serve as a performance reference. Target coverage, coverage uniformity and SNP calling accuracy obtained using any of the WGA, is inferior to the results obtained on bulk cell samples. However, results after REPLI-g come close. Compared to the other WGA methods, the method using REPLI-g WGA results in a better coverage of the targeted genomic regions with a more uniform read depth. Consequently, this method also results in a more accurate SNP calling and could be considered for clinical genotyping of a limited number of cells.
Conflict of interest statement
Figures
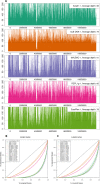
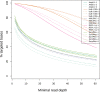
References
-
- Zegers-Hochschild F, Adamson GD, Dyer S, Racowsky C, de Mouzon J, Sokol R, et al. The International Glossary on Infertility and Fertility Care, 2017†‡§. Hum Reprod. 2017. September 1;32(9):1786–801. doi: 10.1093/humrep/dex234 - DOI - PMC - PubMed
-
- Deleye L, Dheedene A, De Coninck D, Sante T, Christodoulou C, Heindryckx B, et al. Shallow whole genome sequencing is well suited for the detection of chromosomal aberrations in human blastocysts. Fertil Steril. 2015. November 1;104(5):1276–1285.e1. doi: 10.1016/j.fertnstert.2015.07.1144 - DOI - PubMed
-
- Fiorentino F, Biricik A, Bono S, Spizzichino L, Cotroneo E, Cottone G, et al. Development and validation of a next-generation sequencing-based protocol for 24-chromosome aneuploidy screening of embryos. Fertil Steril. 2014. May 1;101(5):1375–1382.e2. doi: 10.1016/j.fertnstert.2014.01.051 - DOI - PubMed
-
- Wells D, Kaur K, Grifo J, Glassner M, Taylor JC, Fragouli E, et al. Clinical utilisation of a rapid low-pass whole genome sequencing technique for the diagnosis of aneuploidy in human embryos prior to implantation. J Med Genet. 2014. August 1;51(8):553–62. doi: 10.1136/jmedgenet-2014-102497 - DOI - PMC - PubMed
-
- Dreesen J, Destouni A, Kourlaba G, Degn B, Mette WC, Carvalho F, et al. Evaluation of PCR-based preimplantation genetic diagnosis applied to monogenic diseases: A collaborative ESHRE PGD consortium study. Eur J Hum Genet. 2014. August 4;22(8):1012–8. doi: 10.1038/ejhg.2013.277 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

