Differentially expressed microRNAs associated with changes of transcript levels in detoxification pathways and DDT-resistance in the Drosophila melanogaster strain 91-R
- PMID: 29698530
- PMCID: PMC5919617
- DOI: 10.1371/journal.pone.0196518
Differentially expressed microRNAs associated with changes of transcript levels in detoxification pathways and DDT-resistance in the Drosophila melanogaster strain 91-R
Abstract
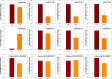
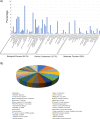
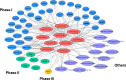
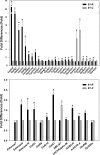
Dichloro-diphenyl-trichloroethane (DDT) resistance among arthropod species is a model for understanding the molecular adaptations in response to insecticide exposures. Previous studies reported that DDT resistance may involve one or multiple detoxification genes, such as cytochrome P450 monooxygenases (P450s), glutathione S-transferases (GSTs), esterases, and ATP binding cassette (ABC) transporters, or changes in the voltage-sensitive sodium channel. However, the possible involvement of microRNAs (miRNAs) in the post-transcriptional regulation of genes associated with DDT resistance in the Drosophila melanogaster strain 91-R remains poorly understood. In this study, the majority of the resulting miRNAs discovered in small RNA libraries from 91-R and the susceptible control strain, 91-C, ranged from 16-25 nt, and contained 163 precursors and 256 mature forms of previously-known miRNAs along with 17 putative novel miRNAs. Quantitative analyses predicted the differential expression of ten miRNAs between 91-R and 91-C, and, based on Gene Ontology and pathway analysis, these ten miRNAs putatively target transcripts encoding proteins involved in detoxification mechanisms. RT-qPCR validated an inverse correlation between levels of differentially-expressed miRNAs and their putatively targeted transcripts, which implies a role of these miRNAs in the differential regulation of detoxification pathways in 91-R compared to 91-C. This study provides evidence associating the differential expression of miRNAs in response to multigenerational DDT selection in Drosophila melanogaster and provides important clues for understanding the possible roles of miRNAs in mediating insecticide resistance traits.
Conflict of interest statement
Figures
References
-
- Casida JE, Quistad GB. Golden age of insecticide research: past, present, or future? Annu Rev Entomol. 1998;43: 1–16. doi: 10.1146/annurev.ento.43.1.1 - DOI - PubMed
-
- Roush RT, McKenzie JA. Ecological genetics of insecticide and acaricide resistance. Annu Rev Entomol. 1987;32: 361–380. doi: 10.1146/annurev.en.32.010187.002045 - DOI - PubMed
-
- Mallet J. The evolution of insecticide resistance: Have the insects won? Trends Ecol Evol. 1989;4: 336–340. doi: 10.1016/0169-5347(89)90088-8 - DOI - PubMed
-
- Hemingway J, Ranson H. Insecticide resistance in insect vectors of human disease. Annu Rev Entomol. 2000;45: 371–391. doi: 10.1146/annurev.ento.45.1.371 - DOI - PubMed
-
- Organization WH. Indoor residual spraying. Use of indoor residual spraying for scaling up global malaria control and elimination. Geneva, Switzerland (WHO/HTM/MAL/20061112); 2006.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

