Precision Profiling of the Cardiovascular Post-Translationally Modified Proteome: Where There Is a Will, There Is a Way
- PMID: 29700069
- PMCID: PMC5963703
- DOI: 10.1161/CIRCRESAHA.118.310966
Precision Profiling of the Cardiovascular Post-Translationally Modified Proteome: Where There Is a Will, There Is a Way
Abstract
There is an exponential increase in biological complexity as initial gene transcripts are spliced, translated into amino acid sequence, and post-translationally modified. Each protein can exist as multiple chemical or sequence-specific proteoforms, and each has the potential to be a critical mediator of a physiological or pathophysiological signaling cascade. Here, we provide an overview of how different proteoforms come about in biological systems and how they are most commonly measured using mass spectrometry-based proteomics and bioinformatics. Our goal is to present this information at a level accessible to every scientist interested in mass spectrometry and its application to proteome profiling. We will specifically discuss recent data linking various protein post-translational modifications to cardiovascular disease and conclude with a discussion for enablement and democratization of proteomics across the cardiovascular and scientific community. The aim is to inform and inspire the readership to explore a larger breadth of proteoform, particularity post-translational modifications, related to their particular areas of expertise in cardiovascular physiology.
Keywords: cardiovascular diseases; mass spectrometry; post-translational protein modifications; proteome; proteomics.
© 2018 American Heart Association, Inc.
Figures
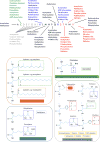

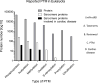

References
-
- Morita H. Human genomics in cardiovascular medicine: implications and perspectives. Circ J. 2013;77:876–85. - PubMed
-
- Aebersold R, Agar JN, Amster IJ, Baker MS, Bertozzi CR, Boja ES, Costello CE, Cravatt BF, Fenselau C, Garcia BA, Ge Y, Gunawardena J, Hendrickson RC, Hergenrother PJ, Huber CG, Ivanov AR, Jensen ON, Jewett MC, Kelleher NL, Kiessling LL, Krogan NJ, Larsen MR, Loo JA, Ogorzalek Loo RR, Lundberg E, MacCoss MJ, Mallick P, Mootha VK, Mrksich M, Muir TW, Patrie SM, Pesavento JJ, Pitteri SJ, Rodriguez H, Saghatelian A, Sandoval W, Schlüter H, Sechi S, Slavoff SA, Smith LM, Snyder MP, Thomas PM, Uhlén M, Van Eyk JE, Vidal M, Walt DR, White FM, Williams ER, Wohlschlager T, Wysocki VH, Yates NA, Young NL, Zhang B. How many human proteoforms are there? Nat Chem Biol. 2018;14:206–214. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

