Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo
- PMID: 29700229
- PMCID: PMC6083445
- DOI: 10.1126/science.aar4362
Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo
Abstract
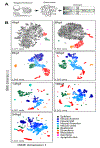
High-throughput mapping of cellular differentiation hierarchies from single-cell data promises to empower systematic interrogations of vertebrate development and disease. Here we applied single-cell RNA sequencing to >92,000 cells from zebrafish embryos during the first day of development. Using a graph-based approach, we mapped a cell-state landscape that describes axis patterning, germ layer formation, and organogenesis. We tested how clonally related cells traverse this landscape by developing a transposon-based barcoding approach (TracerSeq) for reconstructing single-cell lineage histories. Clonally related cells were often restricted by the state landscape, including a case in which two independent lineages converge on similar fates. Cell fates remained restricted to this landscape in embryos lacking the chordin gene. We provide web-based resources for further analysis of the single-cell data.
Copyright © 2018 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures




Comment in
-
A new view of embryo development and regeneration.Science. 2018 Jun 1;360(6392):967-968. doi: 10.1126/science.aat8413. Science. 2018. PMID: 29853675 No abstract available.
References
-
- Hashimshony T, Senderovich N, Avital G, Klochendler A, de Leeuw Y, Anavy L, Gennert D, Li S, Livak KJ, Rozenblatt-Rosen O, Dor Y, Regev A, Yanai I, CEL-Seq2: Sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol. 17, 77 (2016). doi:10.1186/s13059-016-0938-8 10.1186/s13059-016-0938-8MedlineMedline - DOI - PMC - PubMed
-
- Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW, Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201 (2015). doi:10.1016/j.cell.2015.04.044 10.1016/j.cell.2015.04.044MedlineMedline - DOI - PMC - PubMed
-
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA, Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015). doi:10.1016/j.cell.2015.05.002 10.1016/j.cell.2015.05.002MedlineMedline - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

