Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression
- PMID: 29700475
- PMCID: PMC5934326
- DOI: 10.1038/s41588-018-0090-3
Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression
Abstract
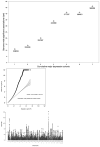
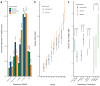
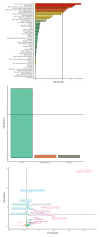
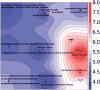
Major depressive disorder (MDD) is a common illness accompanied by considerable morbidity, mortality, costs, and heightened risk of suicide. We conducted a genome-wide association meta-analysis based in 135,458 cases and 344,901 controls and identified 44 independent and significant loci. The genetic findings were associated with clinical features of major depression and implicated brain regions exhibiting anatomical differences in cases. Targets of antidepressant medications and genes involved in gene splicing were enriched for smaller association signal. We found important relationships of genetic risk for major depression with educational attainment, body mass, and schizophrenia: lower educational attainment and higher body mass were putatively causal, whereas major depression and schizophrenia reflected a partly shared biological etiology. All humans carry lesser or greater numbers of genetic risk factors for major depression. These findings help refine the basis of major depression and imply that a continuous measure of risk underlies the clinical phenotype.
Conflict of interest statement
Aartjan TF Beekman: Speakers bureaus of Lundbeck and GlaxoSmithKline. Greg Crawford: Co-founder of Element Genomics. Enrico Domenici: Employee of Hoffmann-La Roche at the time this study was conducted, consultant to Roche and Pierre-Fabre. Nicholas Eriksson: Employed by 23andMe, Inc. and owns stock in 23andMe, Inc. David Hinds: Employee of and own stock options in 23andMe, Inc. Sara Paciga: Employee of Pfizer, Inc. Craig L Hyde: Employee of Pfizer, Inc. Ashley R Winslow: Former employee and stockholder of Pfizer, Inc. Jorge A Quiroz: Employee of Hoffmann-La Roche at the time this study was conducted. Hreinn Stefansson: Employee of deCODE Genetics/AMGEN. Kari Stefansson: Employee of deCODE Genetics/AMGEN. Stacy Steinberg: Employee of deCODE Genetics/AMGEN. Patrick F Sullivan: Scientific advisory board for Pfizer Inc and an advisory committee for Lundbeck. Thorgeir E Thorgeirsson: Employee of deCODE Genetics/AMGEN. Chao Tian: Employee of and own stock options in 23andMe, Inc.
Figures
References
-
- Judd LL. The clinical course of unipolar major depressive disorders. Archives of general psychiatry. 1997;54:989–91. - PubMed
-
- Lopez AD, Mathers CD, Ezzati M, Jamison DT, Murray CJ. Global and regional burden of disease and risk factors, 2001: systematic analysis of population health data. Lancet. 2006;367:1747–57. - PubMed
-
- Wittchen HU, et al. The size and burden of mental disorders and other disorders of the brain in Europe 2010. European neuropsychopharmacology : the journal of the European College of Neuropsychopharmacology. 2011;21:655–79. - PubMed
Publication types
MeSH terms
Grants and funding
- U01 MH085520/MH/NIMH NIH HHS/United States
- P50 CA097007/CA/NCI NIH HHS/United States
- U01 MH046276/MH/NIMH NIH HHS/United States
- MC_QA137853/MRC_/Medical Research Council/United Kingdom
- MR/K026992/1/MRC_/Medical Research Council/United Kingdom
- R01 MH067257/MH/NIMH NIH HHS/United States
- R01 MH059587/MH/NIMH NIH HHS/United States
- MC_PC_U127561128/MRC_/Medical Research Council/United Kingdom
- R01 MH059586/MH/NIMH NIH HHS/United States
- P50 CA093459/CA/NCI NIH HHS/United States
- U01 MH109528/MH/NIMH NIH HHS/United States
- R01 MH060879/MH/NIMH NIH HHS/United States
- R01 MH061675/MH/NIMH NIH HHS/United States
- MC_UU_00007/10/MRC_/Medical Research Council/United Kingdom
- MR/N015746/1/MRC_/Medical Research Council/United Kingdom
- R01 CA133996/CA/NCI NIH HHS/United States
- T32 HL007901/HL/NHLBI NIH HHS/United States
- R01 ES011740/ES/NIEHS NIH HHS/United States
- U01 MH079469/MH/NIMH NIH HHS/United States
- R01 MH060870/MH/NIMH NIH HHS/United States
- R01 MH081800/MH/NIMH NIH HHS/United States
- R01 DA034076/DA/NIDA NIH HHS/United States
- R01 MH059571/MH/NIMH NIH HHS/United States
- G0200243/MRC_/Medical Research Council/United Kingdom
- R01 MH059565/MH/NIMH NIH HHS/United States
- U01 MH109536/MH/NIMH NIH HHS/United States
- U01 MH079470/MH/NIMH NIH HHS/United States
- R01 HG009658/HG/NHGRI NIH HHS/United States
- U01 MH109532/MH/NIMH NIH HHS/United States
- R01 MH059566/MH/NIMH NIH HHS/United States
- K01 MH109772/MH/NIMH NIH HHS/United States
- U01 MH094421/MH/NIMH NIH HHS/United States
- P30 GM103328/GM/NIGMS NIH HHS/United States
- MC_PC_17228/MRC_/Medical Research Council/United Kingdom
- S10 OD018164/OD/NIH HHS/United States
- R01 MH059588/MH/NIMH NIH HHS/United States
- U01 MH046318/MH/NIMH NIH HHS/United States
- MR/L023784/2/MRC_/Medical Research Council/United Kingdom
- MR/L010305/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

