The eXceptional nature of the X chromosome
- PMID: 29701779
- PMCID: PMC6061837
- DOI: 10.1093/hmg/ddy148
The eXceptional nature of the X chromosome
Abstract
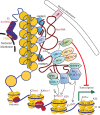
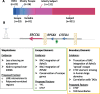
The X chromosome is unique in the genome. In this review we discuss recent advances in our understanding of the genetics and epigenetics of the X chromosome. The X chromosome shares limited conservation with its ancestral homologue the Y chromosome and the resulting difference in X-chromosome dosage between males and females is largely compensated for by X-chromosome inactivation. The process of inactivation is initiated by the long non-coding RNA X-inactive specific transcript (XIST) and achieved through interaction with multiple synergistic silencing pathways. Identification of Xist-interacting proteins has given insight into these processes yet the cascade of events from initiation to maintenance have still to be resolved. In particular, the initiation of inactivation in humans has been challenging to study as: it occurs very early in development; most human embryonic stem cell lines already have an inactive X; and the process seems to differ from mouse. Another difference between human and mouse X inactivation is the larger number of human genes that escape silencing. In humans over 20% of X-linked genes continue to be expressed from the otherwise inactive X chromosome. We are only beginning to understand how such escape occurs but there is growing recognition that escapees contribute to sexually dimorphic traits. The unique biology and epigenetics of the X chromosome have often led to its exclusion from disease studies, yet the X constitutes 5% of the genome and is an important contributor to disease, often in a sex-specific manner.
Figures

References
-
- Hughes J.F., Page D.C. (2016) The history of the Y chromosome in man. Nat. Genet., 48, 588–589. - PubMed
-
- Veitia R.A., Veyrunes F., Bottani S., Birchler J.A. (2015) X chromosome inactivation and active X upregulation in therian mammals: facts, questions, and hypotheses. J. Mol. Cell. Biol., 7, 2–11. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

