One step generation of customizable gRNA vectors for multiplex CRISPR approaches through string assembly gRNA cloning (STAgR)
- PMID: 29702666
- PMCID: PMC5922533
- DOI: 10.1371/journal.pone.0196015
One step generation of customizable gRNA vectors for multiplex CRISPR approaches through string assembly gRNA cloning (STAgR)
Abstract
Novel applications based on the bacterial CRISPR system make genetic, genomic, transcriptional and epigenomic engineering widely accessible for the first time. A significant advantage of CRISPR over previous methods is its tremendous adaptability due to its bipartite nature. Cas9 or its engineered variants define the molecular effect, while short gRNAs determine the targeting sites. A majority of CRISPR approaches depend on the simultaneous delivery of multiple gRNAs into single cells, either as an essential precondition, to increase responsive cell populations or to enhance phenotypic outcomes. Despite these requirements, methods allowing the efficient generation and delivery of multiple gRNA expression units into single cells are still sparse. Here we present STAgR (String assembly gRNA cloning), a single step gRNA multiplexing system, that obtains its advantages by employing the N20 targeting sequences as necessary homologies for Gibson assembly. We show that STAgR allows reliable and cost-effective generation of vectors with high numbers of gRNAs enabling multiplexed CRISPR approaches. Moreover, STAgR is easily customizable, as vector backbones as well as gRNA structures, numbers and promoters can be freely chosen and combined. Finally, we demonstrate STAgR's widespread functionality, its efficiency in multi-targeting approaches, using it for both, genome and transcriptome editing, as well as applying it in vitro and in vivo.
Conflict of interest statement
Figures

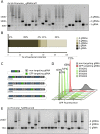
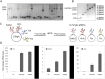
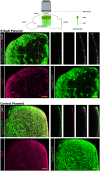
References
-
- Ledford H. CRISPR: gene editing is just the beginning. Nature. 2016;531(7593):156–9. doi: 10.1038/531156a . - DOI - PubMed
-
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–23. doi: 10.1126/science.1231143 - DOI - PMC - PubMed
-
- Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471 doi: 10.7554/eLife.00471 - DOI - PMC - PubMed
-
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–21. doi: 10.1126/science.1225829 . - DOI - PMC - PubMed
-
- Köferle A, Stricker SH, Beck S. Brave new epigenomes: the dawn of epigenetic engineering. Genome Med. 2015;7(1):59 doi: 10.1186/s13073-015-0185-8 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

