A Plasma Biomarker Panel of Four MicroRNAs for the Diagnosis of Prostate Cancer
- PMID: 29703916
- PMCID: PMC5923293
- DOI: 10.1038/s41598-018-24424-w
A Plasma Biomarker Panel of Four MicroRNAs for the Diagnosis of Prostate Cancer
Abstract
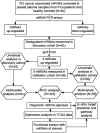
Prostate cancer is diagnosed in over 1 million men every year globally, yet current diagnostic modalities are inadequate for identification of significant cancer and more reliable early diagnostic biomarkers are necessary for improved clinical management of prostate cancer patients. MicroRNAs (miRNAs) modulate important cellular processes/pathways contributing to cancer and are stably present in body fluids. In this study we profiled 372 cancer-associated miRNAs in plasma collected before (~60% patients) and after/during commencement of treatment (~40% patients), from age-matched prostate cancer patients and healthy controls, and observed elevated levels of 4 miRNAs - miR-4289, miR-326, miR-152-3p and miR-98-5p, which were validated in an independent cohort. The miRNA panel was able to differentiate between prostate cancer patients and controls (AUC = 0.88). Analysis of published miRNA transcriptomic data from clinical samples demonstrated low expression of miR-152-3p in tumour compared to adjacent non-malignant tissues. Overexpression of miR-152-3p increased proliferation and migration of prostate cancer cells, suggesting a role for this miRNA in prostate cancer pathogenesis, a concept that was supported by pathway analysis of predicted miR-152-3p target genes. In summary, a four miRNA panel, including miR-152-3p which likely targets genes with key roles in prostate cancer pathogenesis, has the potential to improve early prostate cancer diagnosis.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Rosser, C. J. & Parker, A. Re: The prostate specific antigen era in the United States is over for prostate cancer: what happened in the last 20 years? J Urol174, 1154–1155; author reply 1155–1156, 10.1097/01.ju.0000169211.49050.8f (2005). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

