Transporter gene acquisition and innovation in the evolution of Microsporidia intracellular parasites
- PMID: 29703975
- PMCID: PMC5923384
- DOI: 10.1038/s41467-018-03923-4
Transporter gene acquisition and innovation in the evolution of Microsporidia intracellular parasites
Abstract
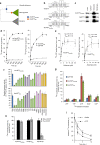
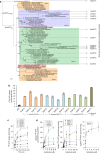
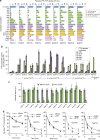
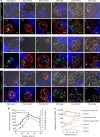
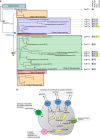
The acquisition of genes by horizontal transfer can impart entirely new biological functions and provide an important route to major evolutionary innovation. Here we have used ancient gene reconstruction and functional assays to investigate the impact of a single horizontally transferred nucleotide transporter into the common ancestor of the Microsporidia, a major radiation of intracellular parasites of animals and humans. We show that this transporter provided early microsporidians with the ability to steal host ATP and to become energy parasites. Gene duplication enabled the diversification of nucleotide transporter function to transport new substrates, including GTP and NAD+, and to evolve the proton-energized net import of nucleotides for nucleic acid biosynthesis, growth and replication. These innovations have allowed the loss of pathways for mitochondrial and cytosolic energy generation and nucleotide biosynthesis that are otherwise essential for free-living eukaryotes, resulting in the highly unusual and reduced cells and genomes of contemporary Microsporidia.
Conflict of interest statement
The authors declare no competing interests.
Figures
Comment in
-
Microsporidia: A Single Horizontal Gene Transfer Drives a Great Leap Forward.Curr Biol. 2018 Jun 18;28(12):R712-R715. doi: 10.1016/j.cub.2018.05.031. Curr Biol. 2018. PMID: 29920267
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

