Low doses of DNA damaging agents extend Saccharomyces cerevisiae chronological lifespan by promoting entry into quiescence
- PMID: 29705357
- PMCID: PMC5994204
- DOI: 10.1016/j.exger.2018.04.020
Low doses of DNA damaging agents extend Saccharomyces cerevisiae chronological lifespan by promoting entry into quiescence
Abstract
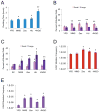
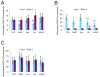
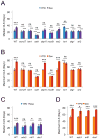
A variety of mild stresses have been shown to extend lifespan in diverse species through hormesis, which is a beneficial response to a stress or toxin that would cause a negative response at a higher exposure. Whether particular stresses induce hormesis can vary with genotype for a given species, and the underlying mechanisms of lifespan extension are only partly understood in most cases. We show that low doses of the DNA damaging or replication stress agents hydroxyurea, methyl methanesulfonate, 4-nitroquinoline 1-oxide, or Zeocin (a phleomycin derivative) lengthened chronological lifespan in Saccharomyces cerevisiae if cells were exposed during growth, but not if they were exposed during stationary phase. Treatment with these agents did not change mitochondrial activity, increase resistance to acetic acid, ethanol, or heat stress, and three of four treatments did not increase resistance to hydrogen peroxide. Stationary phase yeast populations consist of both quiescent and nonquiescent cells, and all four treatments increased the proportion of quiescent cells. Several mutant strains with deletions in genes that influence quiescence prevented Zeocin treatment from extending lifespan and from increasing the proportion of quiescent stationary phase cells. These data indicate that mild DNA damage stress can extend lifespan by promoting quiescence in the absence of mitohormesis or improved general stress responses that have been frequently associated with improved longevity in other cases of hormesis. Further study of the underlying mechanism may yield new insights into quiescence regulation that will be relevant to healthy aging.
Keywords: Aging; Chronological lifespan; DNA damage; Hormesis; Quiescence; Saccharomyces cerevisiae.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Ackerman SH, Tzagoloff A. ATP10, a yeast nuclear gene required for the assembly of the mitochondrial F1-F0 complex. J Biol Chem. 1990;265:9952–9959. - PubMed
-
- Amberg DC, Burke DJ, Strathern JN. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. 2005. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2005.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

