Brain Transcriptomic Analysis of Hereditary Cerebral Hemorrhage With Amyloidosis-Dutch Type
- PMID: 29706885
- PMCID: PMC5908973
- DOI: 10.3389/fnagi.2018.00102
Brain Transcriptomic Analysis of Hereditary Cerebral Hemorrhage With Amyloidosis-Dutch Type
Abstract
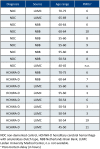
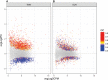
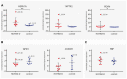
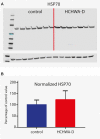
Hereditary cerebral hemorrhage with amyloidosis-Dutch type (HCHWA-D) is an early onset hereditary form of cerebral amyloid angiopathy (CAA) caused by a point mutation resulting in an amino acid change (NP_000475.1:p.Glu693Gln) in the amyloid precursor protein (APP). Post-mortem frontal and occipital cortical brain tissue from nine patients and nine age-related controls was used for RNA sequencing to identify biological pathways affected in HCHWA-D. Although previous studies indicated that pathology is more severe in the occipital lobe in HCHWA-D compared to the frontal lobe, the current study showed similar changes in gene expression in frontal and occipital cortex and the two brain regions were pooled for further analysis. Significantly altered pathways were analyzed using gene set enrichment analysis (GSEA) on 2036 significantly differentially expressed genes. Main pathways over-represented by down-regulated genes were related to cellular aerobic respiration (including ATP synthesis and carbon metabolism) indicating a mitochondrial dysfunction. Principal up-regulated pathways were extracellular matrix (ECM)-receptor interaction and ECM proteoglycans in relation with an increase in the transforming growth factor beta (TGFβ) signaling pathway. Comparison with the publicly available dataset from pre-symptomatic APP-E693Q transgenic mice identified overlap for the ECM-receptor interaction pathway, indicating that ECM modification is an early disease specific pathomechanism.
Keywords: E22Q amyloid β; E693Q mutation; RNA sequencing and transcriptome analysis; extracellular matrix remodeling; familial cerebral amyloid angiopathy; hereditary cerebral hemorrhage with amyloidosis-Dutch type; mitochondrial dysfunction.
Figures




References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

