A Serum Circulating miRNA Signature for Short-Term Risk of Progression to Active Tuberculosis Among Household Contacts
- PMID: 29706954
- PMCID: PMC5908968
- DOI: 10.3389/fimmu.2018.00661
A Serum Circulating miRNA Signature for Short-Term Risk of Progression to Active Tuberculosis Among Household Contacts
Abstract
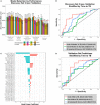
Biomarkers that predict who among recently Mycobacterium tuberculosis (MTB)-exposed individuals will progress to active tuberculosis are urgently needed. Intracellular microRNAs (miRNAs) regulate the host response to MTB and circulating miRNAs (c-miRNAs) have been developed as biomarkers for other diseases. We performed machine-learning analysis of c-miRNA measurements in the serum of adult household contacts (HHCs) of TB index cases from South Africa and Uganda and developed a c-miRNA-based signature of risk for progression to active TB. This c-miRNA-based signature significantly discriminated HHCs within 6 months of progression to active disease from HHCs that remained healthy in an independent test set [ROC area under the ROC curve (AUC) 0.74, progressors < 6 Mo to active TB and ROC AUC 0.66, up to 24 Mo to active TB], and complements the predictions of a previous cellular mRNA-based signature of TB risk.
Keywords: biomarker; correlate of risk; household contact; machine learning; microRNA; tuberculosis.
Figures

References
-
- Marais BJ, Gie RP, Schaaf HS, Hesseling AC, Obihara CC, Starke JJ, et al. The natural history of childhood intra-thoracic tuberculosis: a critical review of literature from the pre-chemotherapy era. Int J Tuberc Lung Dis (2004) 8:392–402. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

