Long Non-Coding RNAs: Novel Players in Regulation of Immune Response Upon Herpesvirus Infection
- PMID: 29706968
- PMCID: PMC5906719
- DOI: 10.3389/fimmu.2018.00761
Long Non-Coding RNAs: Novel Players in Regulation of Immune Response Upon Herpesvirus Infection
Abstract
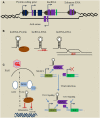
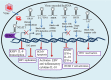
Herpesviruses have developed a variety of sophisticated immune evasion strategies to establish lifelong latent infection, including the use of long non-coding RNAs (lncRNAs). In this review, we summarize the lncRNA action modes, i.e., RNA-protein, RNA-RNA, and RNA-DNA interactions, involved in regulating important aspects of immunity by controlling gene expression at various stages. Upon herpesvirus infection, host lncRNAs, such as nuclear paraspeckle assembly transcript 1, negative regulator of antiviral, and B-cell integration cluster have been functionally characterized as negative or positive antiviral regulators in the immune response. Herpesviruses have also evolved multiple strategies to modulate the host immune response using lncRNAs, such as latency-associated transcript, β 2.7 RNA, 5 kb and 7.2 kb lncRNAs, Epstein-Barr virus-encoded non-coding RNA, BamH I-A rightward transcripts, polyadenylated nuclear, and herpesvirus saimiri U-rich RNAs. We discuss the various mechanisms of immune-related lncRNAs, and their diversified and important functions in the modulation of innate and adaptive immunity upon herpesvirus infection as well as in host-pathogen interactions, which will facilitate our understanding of rational design of novel strategies to combat herpesvirus infection.
Keywords: adaptive immunity; herpesvirus; host–pathogen interaction; innate immunity; long non-coding RNAs; virus infection.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

