Breast MRI radiomics: comparison of computer- and human-extracted imaging phenotypes
- PMID: 29708200
- PMCID: PMC5909355
- DOI: 10.1186/s41747-017-0025-2
Breast MRI radiomics: comparison of computer- and human-extracted imaging phenotypes
Abstract
Background: In this study, we sought to investigate if computer-extracted magnetic resonance imaging (MRI) phenotypes of breast cancer could replicate human-extracted size and Breast Imaging-Reporting and Data System (BI-RADS) imaging phenotypes using MRI data from The Cancer Genome Atlas (TCGA) project of the National Cancer Institute.
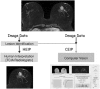
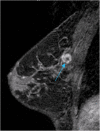
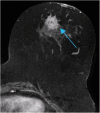
Methods: Our retrospective interpretation study involved analysis of Health Insurance Portability and Accountability Act-compliant breast MRI data from The Cancer Imaging Archive, an open-source database from the TCGA project. This study was exempt from institutional review board approval at Memorial Sloan Kettering Cancer Center and the need for informed consent was waived. Ninety-one pre-operative breast MRIs with verified invasive breast cancers were analysed. Three fellowship-trained breast radiologists evaluated the index cancer in each case according to size and the BI-RADS lexicon for shape, margin, and enhancement (human-extracted image phenotypes [HEIP]). Human inter-observer agreement was analysed by the intra-class correlation coefficient (ICC) for size and Krippendorff's α for other measurements. Quantitative MRI radiomics of computerised three-dimensional segmentations of each cancer generated computer-extracted image phenotypes (CEIP). Spearman's rank correlation coefficients were used to compare HEIP and CEIP.
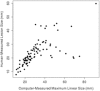
Results: Inter-observer agreement for HEIP varied, with the highest agreement seen for size (ICC 0.679) and shape (ICC 0.527). The computer-extracted maximum linear size replicated the human measurement with p < 10-12. CEIP of shape, specifically sphericity and irregularity, replicated HEIP with both p values < 0.001. CEIP did not demonstrate agreement with HEIP of tumour margin or internal enhancement.
Conclusions: Quantitative radiomics of breast cancer may replicate human-extracted tumour size and BI-RADS imaging phenotypes, thus enabling precision medicine.
Keywords: Breast cancer; Inter-observer variability; Machine learning; Magnetic resonance imaging; Radiomics.
Conflict of interest statement
In this retrospective study, all patient data were Health Insurance Portability and Accountability Act-compliant and acquired under institutional review board approval that waived the need for informed consent.Not applicable.The authors declare that they have no competing interests.Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

References
-
- Morris E, Comstock C, Lee C, et al. ACR BI-RADS® Atlas, Breast Imaging Reporting and Data System. Reston: American College of Radiology; 2013. ACR BI-RADS® magnetic resonance imaging.
-
- Heye T, Merkle EM, Reiner CS, et al. Reproducibility of dynamic contrast-enhanced MR imaging. Part II. Comparison of intra- and interobserver variability with manual region of interest placement versus semiautomatic lesion segmentation and histogram analysis. Radiology. 2013;266:812–821. doi: 10.1148/radiol.12120255. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
