Expanding an expanded genome: long-read sequencing of Trypanosoma cruzi
- PMID: 29708484
- PMCID: PMC5994713
- DOI: 10.1099/mgen.0.000177
Expanding an expanded genome: long-read sequencing of Trypanosoma cruzi
Abstract
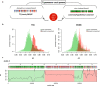
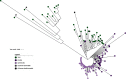
Although the genome of Trypanosoma cruzi, the causative agent of Chagas disease, was first made available in 2005, with additional strains reported later, the intrinsic genome complexity of this parasite (the abundance of repetitive sequences and genes organized in tandem) has traditionally hindered high-quality genome assembly and annotation. This also limits diverse types of analyses that require high degrees of precision. Long reads generated by third-generation sequencing technologies are particularly suitable to address the challenges associated with T. cruzi's genome since they permit direct determination of the full sequence of large clusters of repetitive sequences without collapsing them. This, in turn, not only allows accurate estimation of gene copy numbers but also circumvents assembly fragmentation. Here, we present the analysis of the genome sequences of two T. cruzi clones: the hybrid TCC (TcVI) and the non-hybrid Dm28c (TcI), determined by PacBio Single Molecular Real-Time (SMRT) technology. The improved assemblies herein obtained permitted us to accurately estimate gene copy numbers, abundance and distribution of repetitive sequences (including satellites and retroelements). We found that the genome of T. cruzi is composed of a 'core compartment' and a 'disruptive compartment' which exhibit opposite GC content and gene composition. Novel tandem and dispersed repetitive sequences were identified, including some located inside coding sequences. Additionally, homologous chromosomes were separately assembled, allowing us to retrieve haplotypes as separate contigs instead of a unique mosaic sequence. Finally, manual annotation of surface multigene families, mucins and trans-sialidases allows now a better overview of these complex groups of genes.
Keywords: Chagas disease; PacBio; Trypanosoma cruzi; whole genome sequencing.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

