Histone acetyltransferase TGF-1 regulates Trichoderma atroviride secondary metabolism and mycoparasitism
- PMID: 29708970
- PMCID: PMC5927414
- DOI: 10.1371/journal.pone.0193872
Histone acetyltransferase TGF-1 regulates Trichoderma atroviride secondary metabolism and mycoparasitism
Abstract
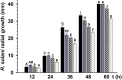
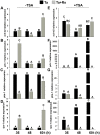
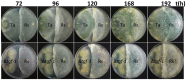
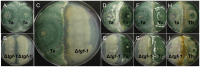
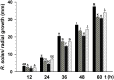
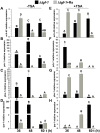
Some filamentous fungi of the Trichoderma genus are used as biocontrol agents against airborne and soilborne phytopathogens. The proposed mechanism by which Trichoderma spp. antagonizes phytopathogens is through the release of lytic enzymes, antimicrobial compounds, mycoparasitism, and the induction of systemic disease-resistance in plants. Here we analyzed the role of TGF-1 (Trichoderma Gcn Five-1), a histone acetyltransferase of Trichoderma atroviride, in mycoparasitism and antibiosis against the phytopathogen Rhizoctonia solani. Trichostatin A (TSA), a histone deacetylase inhibitor that promotes histone acetylation, slightly affected T. atroviride and R. solani growth, but not the growth of the mycoparasite over R. solani. Application of TSA to the liquid medium induced synthesis of antimicrobial compounds. Expression analysis of the mycoparasitism-related genes ech-42 and prb-1, which encode an endochitinase and a proteinase, as well as the secondary metabolism-related genes pbs-1 and tps-1, which encode a peptaibol synthetase and a terpene synthase, respectively, showed that they were regulated by TSA. A T. atroviride strain harboring a deletion of tgf-1 gene showed slow growth, thinner and less branched hyphae than the wild-type strain, whereas its ability to coil around the R. solani hyphae was not affected. Δtgf-1 presented a diminished capacity to grow over R. solani, but the ability of its mycelium -free culture filtrates (MFCF) to inhibit the phytopathogen growth was enhanced. Intriguingly, addition of TSA to the culture medium reverted the enhanced inhibition growth of Δtgf-1 MFCF on R. solani at levels compared to the wild-type MFCF grown in medium amended with TSA. The presence of R. solani mycelium in the culture medium induced similar proteinase activity in a Δtgf-1 compared to the wild-type, whereas the chitinolytic activity was higher in a Δtgf-1 mutant in the absence of R. solani, compared to the parental strain. Expression of mycoparasitism- and secondary metabolism-related genes in Δtgf-1 was differentially regulated in the presence or absence of R. solani. These results indicate that histone acetylation may play important roles in the biocontrol mechanisms of T. atroviride.
Conflict of interest statement
Figures




References
-
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389: 251–60. doi: 10.1038/38444 - DOI - PubMed
-
- He H, Lehming N. Global effects of histone modifications. Brief Funct Genomic Proteomic. 2003;2: 234–43. Available: http://www.ncbi.nlm.nih.gov/pubmed/15239926 - PubMed
-
- Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389: 349–352. doi: 10.1038/38664 - DOI - PubMed
-
- Shahbazian MD, Grunstein M. Functions of Site-Specific Histone Acetylation and Deacetylation. Annu Rev Biochem. 2007;76: 75–100. doi: 10.1146/annurev.biochem.76.052705.162114 - DOI - PubMed
-
- Grimaldi B, Coiro P, Filetici P, Berge E, Dobosy JR, Freitag M, Selker EU B P. The Neurospora crassa White Collar-1 dependent Blue Light Response Requires Acetylation of Histone H3 Lysine 14 by NGF-1. Mol Biol Cell. 2006;17: 4576–4583. doi: 10.1091/mbc.E06-03-0232 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

