Structural and functional dissection of differentially expressed tomato WRKY transcripts in host defense response against the vascular wilt pathogen (Fusarium oxysporum f. sp. lycopersici)
- PMID: 29709017
- PMCID: PMC5927432
- DOI: 10.1371/journal.pone.0193922
Structural and functional dissection of differentially expressed tomato WRKY transcripts in host defense response against the vascular wilt pathogen (Fusarium oxysporum f. sp. lycopersici)
Abstract
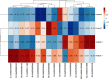
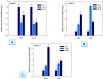
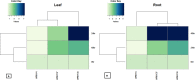
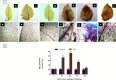
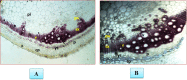
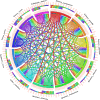
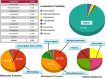
The WRKY transcription factors have indispensable role in plant growth, development and defense responses. The differential expression of WRKY genes following the stress conditions has been well demonstrated. We investigated the temporal and tissue-specific (root and leaf tissues) differential expression of plant defense-related WRKY genes, following the infection of Fusarium oxysporum f. sp. lycopersici (Fol) in tomato. The genome-wide computational analysis revealed that during the Fol infection in tomato, 16 different members of WRKY gene superfamily were found to be involved, of which only three WRKYs (SolyWRKY4, SolyWRKY33, and SolyWRKY37) were shown to have clear-cut differential gene expression. The quantitative real time PCR (qRT-PCR) studies revealed different gene expression profile changes in tomato root and leaf tissues. In root tissues, infected with Fol, an increased expression for SolyWRKY33 (2.76 fold) followed by SolyWRKY37 (1.93 fold) gene was found at 24 hrs which further increased at 48 hrs (5.0 fold). In contrast, the leaf tissues, the expression was more pronounced at an earlier stage of infection (24 hrs). However, in both cases, we found repression of SolyWRKY4 gene, which further decreased at an increased time interval. The biochemical defense programming against Fol pathogenesis was characterized by the highest accumulation of H2O2 (at 48 hrs) and enhanced lignification. The functional diversity across the characterized WRKYs was explored through motif scanning using MEME suite, and the WRKYs specific gene regulation was assessed through the DNA protein docking studies The functional WRKY domain modeled had β sheets like topology with coil and turns. The DNA-protein interaction results revealed the importance of core residues (Tyr, Arg, and Lys) in a feasible WRKY-W-box DNA interaction. The protein interaction network analysis revealed that the SolyWRKY33 could interact with other proteins, such as mitogen-activated protein kinase 5 (MAPK), sigma factor binding protein1 (SIB1) and with other WRKY members including WRKY70, WRKY1, and WRKY40, to respond various biotic and abiotic stresses. The STRING results were further validated through Predicted Tomato Interactome Resource (PTIR) database. The CELLO2GO web server revealed the functional gene ontology annotation and protein subcellular localization, which predicted that SolyWRKY33 is involved in amelioration of biological stress (39.3%) and other metabolic processes (39.3%). The protein (SolyWRKY33) most probably located inside the nucleus (91.3%) with having transcription factor binding activity. We conclude that the defense response following the Fol challenge was accompanied by differential expression of the SolyWRKY4(↓), SolyWRKY33(↑) and SolyWRKY37(↑) transcripts. The biochemical changes are occupied by elicitation of H2O2 generation and accumulation and enhanced lignified tissues.
Conflict of interest statement
Figures








References
-
- Rongai D, Pulcini P, Pesce B, Milano F. Antifungal activity of pomegranate peel extract against fusarium wilt of tomato. Eur J Plant Pathol. 2017; 147:229–238.
-
- Asha BB, Nayaka CS, Shankar UA, Srinivas C, Niranjana SR. Biological control of F. oxysporum f. sp. lycopersici causing wilt of tomato by Pseudomonas fluorescens. Int J Microbial Res. 2011; 3:79.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

