Cancer CRISPR Screens In Vivo
- PMID: 29709259
- PMCID: PMC5935117
- DOI: 10.1016/j.trecan.2018.03.002
Cancer CRISPR Screens In Vivo
Abstract
Clustered regularly interspaced short palindromic repeats (CRISPR) screening is a powerful toolset for investigating diverse biological processes. Most CRISPR screens to date have been performed with in vitro cultures or cellular transplant models. To interrogate cancer in animal models that more closely recapitulate the human disease, autochthonous direct in vivo CRISPR screens have recently been developed that can identify causative drivers in the native tissue microenvironment. By empowering multiplexed mutagenesis in fully immunocompetent animals, direct in vivo CRISPR screens enable the rapid generation of patient-specific avatars that can guide precision medicine. This Opinion article discusses the current status of in vivo CRISPR screens in cancer and offers perspectives on future applications.
Keywords: CRISPR screen; cancer; functional genomics; in vivo.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors have no conflicts of interest related to this review.
Figures
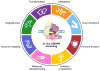
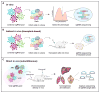

References
-
- Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153:17–37. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

