Amazing Diversity in Biochemical Roles of Fe(II)/2-Oxoglutarate Oxygenases
- PMID: 29709390
- PMCID: PMC6014900
- DOI: 10.1016/j.tibs.2018.04.002
Amazing Diversity in Biochemical Roles of Fe(II)/2-Oxoglutarate Oxygenases
Abstract
Since their discovery in the 1960s, the family of Fe(II)/2-oxoglutarate-dependent oxygenases has undergone a tremendous expansion to include enzymes catalyzing a vast diversity of biologically important reactions. Recent examples highlight roles in controlling chromatin modification, transcription, mRNA demethylation, and mRNA splicing. Others generate modifications in tRNA, translation factors, ribosomes, and other proteins. Thus, oxygenases affect all components of molecular biology's central dogma, in which information flows from DNA to RNA to proteins. These enzymes also function in biosynthesis and catabolism of cellular metabolites, including antibiotics and signaling molecules. Due to their critical importance, ongoing efforts have targeted family members for the development of specific therapeutics. This review provides a general overview of recently characterized oxygenase reactions and their key biological roles.
Keywords: Nonheme iron oxygenase; biodegradation; biosynthesis; chromatin modification; transcription; translation.
Copyright © 2018 Elsevier Ltd. All rights reserved.
Figures


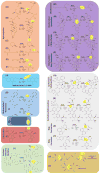
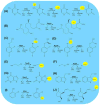
References
-
- Hausinger RP. Biochemical diversity of 2-oxoglutarate-dependent oxygenases. In: Schofield CJ, Hausinger RP, editors. 2-Oxoglutarate-Dependent Oxygenases. Royal Society of Chemistry; 2015. pp. 1–58.
-
- Wu LF, et al. Ferrous iron and α-ketoglutarate dependent dioxygenases in the biosynthesis of microbial natural products. Biochim Biophys Acta. 2016;1864:453–470. - PubMed
-
- Islam MS, et al. 2-Oxoglutarate-dependent oxygenases. Annu Rev Biochem. 2018 in press. - PubMed
-
- Hagel JM, Facchini PJ. Expanding the roles for 2-oxoglutarate-dependent-oxygenases in plant metabolism. Natural Prod Rep. 2018 in press. - PubMed
-
- Aik WS, et al. Introduction to structural studies on 2-oxoglutarate-dependent oxygenases and related enzymes. In: Schofield CJ, Hausinger RP, editors. 2-Oxoglutarate-Dependent Oxygenases. Royal Society of Chemistry; 2015.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

