Remote Memory and Cortical Synaptic Plasticity Require Neuronal CCCTC-Binding Factor (CTCF)
- PMID: 29712785
- PMCID: PMC6705941
- DOI: 10.1523/JNEUROSCI.2738-17.2018
Remote Memory and Cortical Synaptic Plasticity Require Neuronal CCCTC-Binding Factor (CTCF)
Abstract
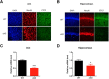
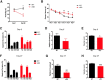
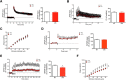
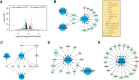
The molecular mechanism of long-term memory has been extensively studied in the context of the hippocampus-dependent recent memory examined within several days. However, months-old remote memory maintained in the cortex for long-term has not been investigated much at the molecular level yet. Various epigenetic mechanisms are known to be important for long-term memory, but how the 3D chromatin architecture and its regulator molecules contribute to neuronal plasticity and systems consolidation is still largely unknown. CCCTC-binding factor (CTCF) is an 11-zinc finger protein well known for its role as a genome architecture molecule. Male conditional knock-out mice in which CTCF is lost in excitatory neurons during adulthood showed normal recent memory in the contextual fear conditioning and spatial water maze tasks. However, they showed remarkable impairments in remote memory in both tasks. Underlying the remote memory-specific phenotypes, we observed that female CTCF conditional knock-out mice exhibit disrupted cortical LTP, but not hippocampal LTP. Similarly, we observed that CTCF deletion in inhibitory neurons caused partial impairment of remote memory. Through RNA sequencing, we observed that CTCF knockdown in cortical neuron culture caused altered expression of genes that are highly involved in cell adhesion, synaptic plasticity, and memory. These results suggest that remote memory storage in the cortex requires CTCF-mediated gene regulation in neurons, whereas recent memory formation in the hippocampus does not.SIGNIFICANCE STATEMENT CCCTC-binding factor (CTCF) is a well-known 3D genome architectural protein that regulates gene expression. Here, we use two different CTCF conditional knock-out mouse lines and reveal, for the first time, that CTCF is critically involved in the regulation of remote memory. We also show that CTCF is necessary for appropriate expression of genes, many of which we found to be involved in the learning- and memory-related processes. Our study provides behavioral and physiological evidence for the involvement of CTCF-mediated gene regulation in the remote long-term memory and elucidates our understanding of systems consolidation mechanisms.
Keywords: 3D genome architecture; CTCF; cortical plasticity; remote memory; systems consolidation.
Copyright © 2018 the authors 0270-6474/18/385042-11$15.00/0.
Figures


Comment in
-
Neuronal Chromatin Architecture Regulator CTCF Dictates Remote Memory.J Neurosci. 2018 Nov 28;38(48):10239-10240. doi: 10.1523/JNEUROSCI.1869-18.2018. J Neurosci. 2018. PMID: 30487296 Free PMC article. No abstract available.
References
-
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet 25:25–29. 10.1038/75556 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
