Galactic Cosmic Radiation Induces Persistent Epigenome Alterations Relevant to Human Lung Cancer
- PMID: 29712937
- PMCID: PMC5928241
- DOI: 10.1038/s41598-018-24755-8
Galactic Cosmic Radiation Induces Persistent Epigenome Alterations Relevant to Human Lung Cancer
Abstract
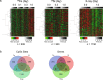
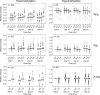
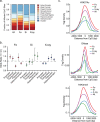
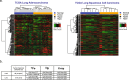
Human deep space and planetary travel is limited by uncertainties regarding the health risks associated with exposure to galactic cosmic radiation (GCR), and in particular the high linear energy transfer (LET), heavy ion component. Here we assessed the impact of two high-LET ions 56Fe and 28Si, and low-LET X rays on genome-wide methylation patterns in human bronchial epithelial cells. We found that all three radiation types induced rapid and stable changes in DNA methylation but at distinct subsets of CpG sites affecting different chromatin compartments. The 56Fe ions induced mostly hypermethylation, and primarily affected sites in open chromatin regions including enhancers, promoters and the edges ("shores") of CpG islands. The 28Si ion-exposure had mixed effects, inducing both hyper and hypomethylation and affecting sites in more repressed heterochromatic environments, whereas X rays induced mostly hypomethylation, primarily at sites in gene bodies and intergenic regions. Significantly, the methylation status of 56Fe ion sensitive sites, but not those affected by X ray or 28Si ions, discriminated tumor from normal tissue for human lung adenocarcinomas and squamous cell carcinomas. Thus, high-LET radiation exposure leaves a lasting imprint on the epigenome, and affects sites relevant to human lung cancer. These methylation signatures may prove useful in monitoring the cumulative biological impact and associated cancer risks encountered by astronauts in deep space.
Conflict of interest statement
The authors declare no competing interests.
Figures


Comment in
-
Epigenetic modification by galactic cosmic radiation as a risk factor for lung cancer: real world data issues.Transl Lung Cancer Res. 2019 Apr;8(2):116-118. doi: 10.21037/tlcr.2019.01.01. Transl Lung Cancer Res. 2019. PMID: 31106121 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

